Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in gamma-hexachlorocyclohexane degradation
- PMID: 16963556
- PMCID: PMC1636184
- DOI: 10.1128/AEM.01531-06
Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in gamma-hexachlorocyclohexane degradation
Abstract
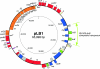
The alpha-proteobacterial strain Sphingobium japonicum UT26 utilizes a highly chlorinated pesticide, gamma-hexachlorocyclohexane (gamma-HCH), as a sole source of carbon and energy, and haloalkane dehalogenase LinB catalyzes the second step of gamma-HCH degradation in UT26. Functional complementation of a linB mutant of UT26, UT26DB, was performed by the exogenous plasmid isolation technique using HCH-contaminated soil, leading to our successful identification of a plasmid, pLB1, carrying the linB gene. Complete sequencing analysis of pLB1, with a size of 65,998 bp, revealed that it carries (i) 50 totally annotated coding sequences, (ii) an IS6100 composite transposon containing two copies of linB, and (iii) potential genes for replication, maintenance, and conjugative transfer with low levels of similarity to other homologues. A minireplicon assay demonstrated that a 2-kb region containing the predicted repA gene and its upstream region of pLB1 functions as an autonomously replicating unit in UT26. Furthermore, pLB1 was conjugally transferred from UT26DB to other alpha-proteobacterial strains but not to any of the beta- or gamma-proteobacterial strains examined to date. These results suggest that this exogenously isolated novel plasmid contributes to the dissemination of at least some genes for gamma-HCH degradation in the natural environment. To the best of our knowledge, this is the first detailed report of a plasmid involved in gamma-HCH degradation.
Figures



Similar articles
-
Degradation of beta-hexachlorocyclohexane by haloalkane dehalogenase LinB from gamma-hexachlorocyclohexane-utilizing bacterium Sphingobium sp. MI1205.Arch Microbiol. 2007 Oct;188(4):313-25. doi: 10.1007/s00203-007-0251-8. Epub 2007 May 22. Arch Microbiol. 2007. PMID: 17516046
-
Isolation and characterization of a novel gamma-hexachlorocyclohexane-degrading bacterium.J Bacteriol. 1996 Oct;178(20):6049-55. doi: 10.1128/jb.178.20.6049-6055.1996. J Bacteriol. 1996. PMID: 8830705 Free PMC article.
-
A novel pathway for the biodegradation of gamma-hexachlorocyclohexane by a Xanthomonas sp. strain ICH12.J Appl Microbiol. 2007 Jun;102(6):1468-78. doi: 10.1111/j.1365-2672.2006.03209.x. J Appl Microbiol. 2007. PMID: 17578411
-
Aerobic degradation of lindane (gamma-hexachlorocyclohexane) in bacteria and its biochemical and molecular basis.Appl Microbiol Biotechnol. 2007 Sep;76(4):741-52. doi: 10.1007/s00253-007-1066-x. Epub 2007 Jul 19. Appl Microbiol Biotechnol. 2007. PMID: 17634937 Review.
-
Comprehensive review on Haloalkane dehalogenase (LinB): a β-hexachlorocyclohexane (HCH) degrading enzyme.Arch Microbiol. 2024 Aug 14;206(9):380. doi: 10.1007/s00203-024-04105-1. Arch Microbiol. 2024. PMID: 39143366 Review.
Cited by
-
Insights into Ongoing Evolution of the Hexachlorocyclohexane Catabolic Pathway from Comparative Genomics of Ten Sphingomonadaceae Strains.G3 (Bethesda). 2015 Apr 7;5(6):1081-94. doi: 10.1534/g3.114.015933. G3 (Bethesda). 2015. PMID: 25850427 Free PMC article.
-
Exogenous isolation of conjugative plasmids from pesticide contaminated soil.World J Microbiol Biotechnol. 2012 Feb;28(2):567-74. doi: 10.1007/s11274-011-0849-5. Epub 2011 Jul 27. World J Microbiol Biotechnol. 2012. PMID: 22806852
-
Nucleotide sequence of plasmid pCNB1 from comamonas strain CNB-1 reveals novel genetic organization and evolution for 4-chloronitrobenzene degradation.Appl Environ Microbiol. 2007 Jul;73(14):4477-83. doi: 10.1128/AEM.00616-07. Epub 2007 May 25. Appl Environ Microbiol. 2007. PMID: 17526790 Free PMC article.
-
Complete nucleotide sequence of TOL plasmid pDK1 provides evidence for evolutionary history of IncP-7 catabolic plasmids.J Bacteriol. 2010 Sep;192(17):4337-47. doi: 10.1128/JB.00359-10. Epub 2010 Jun 25. J Bacteriol. 2010. PMID: 20581207 Free PMC article.
-
Molecular Basis and Evolutionary Origin of 1-Nitronaphthalene Catabolism in Sphingobium sp. Strain JS3065.Appl Environ Microbiol. 2023 Jan 31;89(1):e0172822. doi: 10.1128/aem.01728-22. Epub 2023 Jan 9. Appl Environ Microbiol. 2023. PMID: 36622195 Free PMC article.
References
-
- Bagdasarian, M., R. Lurz, B. Ruckert, F. C. Franklin, M. M. Bagdasarian, J. Frey, and K. N. Timmis. 1981. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237-247. - PubMed
-
- Bale, M. J., J. C. Fry, and M. J. Day. 1987. Plasmid transfer between strains of Pseudomonas aeruginosa on membrane filters attached to river stones. J. Gen. Microbiol. 133:3099-3107. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

