Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae
- PMID: 16963631
- PMCID: PMC1563586
- DOI: 10.1128/EC.00107-06
Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae
Abstract
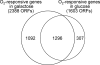
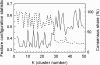
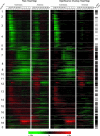
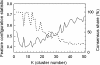
We conducted a comprehensive genomic analysis of the temporal response of yeast to anaerobiosis (six generations) and subsequent aerobic recovery ( approximately 2 generations) to reveal metabolic-state (galactose versus glucose)-dependent differences in gene network activity and function. Analysis of variance showed that far fewer genes responded (raw P value of <or=10(-8)) to the O(2) shifts in glucose (1,603 genes) than in galactose (2,388 genes). Gene network analysis reveals that this difference is due largely to the failure of "stress"-activated networks controlled by Msn2/4, Fhl1, MCB, SCB, PAC, and RRPE to transiently respond to the shift to anaerobiosis in glucose as they did in galactose. After approximately 1 generation of anaerobiosis, the response was similar in both media, beginning with the deactivation of Hap1 and Hap2/3/4/5 networks involved in mitochondrial functions and the concomitant derepression of Rox1-regulated networks for carbohydrate catabolism and redox regulation and ending (>or=2 generations) with the activation of Upc2- and Mot3-regulated networks involved in sterol and cell wall homeostasis. The response to reoxygenation was rapid (<5 min) and similar in both media, dominated by Yap1 networks involved in oxidative stress/redox regulation and the concomitant activation of heme-regulated ones. Our analyses revealed extensive networks of genes subject to combinatorial regulation by both heme-dependent (e.g., Hap1, Hap2/3/4/5, Rox1, Mot3, and Upc2) and heme-independent (e.g., Yap1, Skn7, and Puf3) factors under these conditions. We also uncover novel functions for several cis-regulatory sites and trans-acting factors and define functional regulons involved in the physiological acclimatization to changes in oxygen availability.
Figures


References
-
- Andreasen, A., and T. Stier. 1953. Anaerobic nutrition of Saccharomyces cerevisiae. I. Ergosterol requirement for growth in a defined medium. J. Cell. Comp. Physiol. 41:23-36. - PubMed
-
- Andreasen, A., and T. Stier. 1954. Anaerobic nutrition of Saccharomyces cerevisiae. II. Unsaturated fatty acid requirement for growth in defined medium. J. Cell. Comp. Physiol. 43:271-281. - PubMed
-
- Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

