Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements
- PMID: 16963634
- PMCID: PMC1563581
- DOI: 10.1128/EC.00106-06
Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements
Abstract
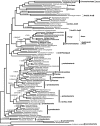
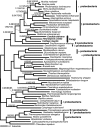
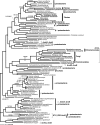
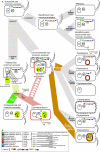
Currently the shikimate pathway is reported as a metabolic feature of prokaryotes, ascomycete fungi, apicomplexans, and plants. The plant shikimate pathway enzymes have similarities to prokaryote homologues and are largely active in chloroplasts, suggesting ancestry from the plastid progenitor genome. Toxoplasma gondii, which also possesses an alga-derived plastid organelle, encodes a shikimate pathway with similarities to ascomycete genes, including a five-enzyme pentafunctional arom. These data suggests that the shikimate pathway and the pentafunctional arom either had an ancient origin in the eukaryotes or was conveyed by eukaryote-to-eukaryote horizontal gene transfer (HGT). We expand sampling and analyses of the shikimate pathway genes to include the oomycetes, ciliates, diatoms, basidiomycetes, zygomycetes, and the green and red algae. Sequencing of cDNA from Tetrahymena thermophila confirmed the presence of a pentafused arom, as in fungi and T. gondii. Phylogenies and taxon distribution suggest that the arom gene fusion event may be an ancient eukaryotic innovation. Conversely, the Plantae lineage (represented here by both Viridaeplantae and the red algae) acquired different prokaryotic genes for all seven steps of the shikimate pathway. Two of the phylogenies suggest a derivation of the Plantae genes from the cyanobacterial plastid progenitor genome, but if the full Plantae pathway was originally of cyanobacterial origin, then the five other shikimate pathway genes were obtained from a minimum of two other eubacterial genomes. Thus, the phylogenies demonstrate both separate HGTs and shared derived HGTs within the Plantae clade either by primary HGT transfer or secondarily via the plastid progenitor genome. The shared derived characters support the holophyly of the Plantae lineage and a single ancestral primary plastid endosymbiosis. Our analyses also pinpoints a minimum of 50 gene/domain loss events, demonstrating that loss and replacement events have been an important process in eukaryote genome evolution.
Figures




References
-
- Andersson, J. O., S. W. Sarchfield, and A. J. Roger. 2005. Gene transfers from nanoarchaeota to an ancestor of diplomonads and parabasalids. Mol. Biol. Evol. 22:85-90. - PubMed
-
- Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. - PubMed
-
- Boucher, Y., C. J. Douady, R. T. Papke, D. A. Walsh, M. E. Boudreau, C. L. Nesbo, R. J. Case, and W. F. Doolittle. 2003. Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37:283-328. - PubMed
-
- Brinkman, F. S., J. L. Blanchard, A. Cherkasov, Y. Av-Gay, R. C. Brunham, R. C. Fernandez, B. B. Finlay, S. P. Otto, B. F. Ouellette, P. J. Keeling, A. M. Rose, R. E. Hancock, S. J. Jones, and H. Greberg. 2002. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 12:1159-1167. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

