Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice
- PMID: 16963705
- PMCID: PMC1581435
- DOI: 10.1101/gr.5290206
Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice
Erratum in
- Genome Res. 2011 Jul;21(7):1201. Saniyal, Abhijit [corrected to Sanyal, Abhijit]
Abstract
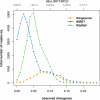
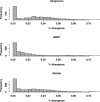
Retrotransposons are the main components of eukaryotic genomes, representing up to 80% of some large plant genomes. These mobile elements transpose via a "copy and paste" mechanism, thus increasing their copy number while active. Their accumulation is now accepted as the main factor of genome size increase in higher eukaryotes, besides polyploidy. However, the dynamics of this process are poorly understood. In this study, we show that Oryza australiensis, a wild relative of the Asian cultivated rice O. sativa, has undergone recent bursts of three LTR-retrotransposon families. This genome has accumulated more than 90,000 retrotransposon copies during the last three million years, leading to a rapid twofold increase of its size. In addition, phenetic analyses of these retrotransposons clearly confirm that the genomic bursts occurred posterior to the radiation of the species. This provides direct evidence of retrotransposon-mediated variation of genome size within a plant genus.
Figures



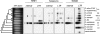

References
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Ammiraju J.S., Luo M., Goicoechea J.L., Wang W., Kudrna D., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., Luo M., Goicoechea J.L., Wang W., Kudrna D., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., Goicoechea J.L., Wang W., Kudrna D., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., Wang W., Kudrna D., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., Kudrna D., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., Mueller C., Talag J., Kim H., Sisneros N.B., Blackmon B., Talag J., Kim H., Sisneros N.B., Blackmon B., Kim H., Sisneros N.B., Blackmon B., Sisneros N.B., Blackmon B., Blackmon B., et al. The Oryza bacterial artificial chromosome library resource: Construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza . Genome Res. 2006;16:140–147. - PMC - PubMed
-
- Bartosch B., Stefanidis D., Myers R., Weiss R., Patience C., Takeuchi Y., Stefanidis D., Myers R., Weiss R., Patience C., Takeuchi Y., Myers R., Weiss R., Patience C., Takeuchi Y., Weiss R., Patience C., Takeuchi Y., Patience C., Takeuchi Y., Takeuchi Y. Evidence and consequence of porcine endogenous retrovirus recombination. J. Virol. 2004;78:13880–13890. - PMC - PubMed
-
- Brar D.S., Khush G.S., Khush G.S. Alien introgression in rice. Plant Mol. Biol. 1997;35:35–47. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
