Kinetic and structural insight into the mechanism of BphD, a C-C bond hydrolase from the biphenyl degradation pathway
- PMID: 16964968
- PMCID: PMC2519953
- DOI: 10.1021/bi0611098
Kinetic and structural insight into the mechanism of BphD, a C-C bond hydrolase from the biphenyl degradation pathway
Abstract
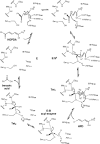
Kinetic and structural analyses of 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (HOPDA) hydrolase from Burkholderia xenovorans LB400 (BphD(LB400)) provide insight into the catalytic mechanism of this unusual serine hydrolase. Single turnover stopped-flow analysis at 25 degrees C showed that the enzyme rapidly (1/tau(1) approximately 500 s(-1)) transforms HOPDA (lambda(max) = 434 nm) into a species with electronic absorption maxima at 473 and 492 nm. The absorbance of this enzyme-bound species (E:S) decayed in a biphasic manner (1/tau(2) = 54 s(-1), 1/tau(3) = 6 s(-1) approximately k(cat)) with simultaneous biphasic appearance (48 and 8 s(-1)) of an absorbance band at 270 nm characteristic of one of the products, 2-hydroxypenta-2,4-dienoic acid (HPD). Increasing solution viscosity with glycerol slowed 1/tau(1) and 1/tau(2) but affected neither 1/tau(3) nor k(cat), suggesting that 1/tau(2) may reflect diffusive HPD dissociation, and 1/tau(3) represents an intramolecular event. Product inhibition studies suggested that the other product, benzoate, is released after HPD. Contrary to studies in a related hydrolase, we found no evidence that ketonized HOPDA is partially released prior to hydrolysis, and, therefore, postulate that the biphasic kinetics reflect one of two mechanisms, pending assignment of E:S (lambda(max) = 492 nm). The crystal structures of the wild type, the S112C variant, and S112C incubated with HOPDA were each determined to 1.6 A resolution. The latter reveals interactions between conserved active site residues and the dienoate moiety of the substrate. Most notably, the catalytic residue His265 is hydrogen-bonded to the 2-hydroxy/oxo substituent of HOPDA, consistent with a role in catalyzing ketonization. The data are more consistent with an acyl-enzyme mechanism than with the formation of a gem-diol intermediate.
Figures





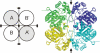
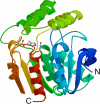


Similar articles
-
The tautomeric half-reaction of BphD, a C-C bond hydrolase. Kinetic and structural evidence supporting a key role for histidine 265 of the catalytic triad.J Biol Chem. 2007 Jul 6;282(27):19894-904. doi: 10.1074/jbc.M702237200. Epub 2007 Apr 18. J Biol Chem. 2007. PMID: 17442675
-
The molecular basis for inhibition of BphD, a C-C bond hydrolase involved in polychlorinated biphenyls degradation: large 3-substituents prevent tautomerization.J Biol Chem. 2007 Dec 14;282(50):36377-85. doi: 10.1074/jbc.M707035200. Epub 2007 Oct 11. J Biol Chem. 2007. PMID: 17932031
-
The catalytic serine of meta-cleavage product hydrolases is activated differently for C-O bond cleavage than for C-C bond cleavage.Biochemistry. 2012 Jul 24;51(29):5831-40. doi: 10.1021/bi300663r. Epub 2012 Jul 11. Biochemistry. 2012. PMID: 22747426
-
The lid domain of the MCP hydrolase DxnB2 contributes to the reactivity toward recalcitrant PCB metabolites.Biochemistry. 2013 Aug 20;52(33):5685-5695. doi: 10.1021/bi400774m. Epub 2013 Aug 9. Biochemistry. 2013. PMID: 23879719 Free PMC article.
-
A product analog bound form of 3-oxoadipate-enol-lactonase (PcaD) reveals a multifunctional role for the divergent cap domain.J Mol Biol. 2011 Mar 11;406(5):649-58. doi: 10.1016/j.jmb.2011.01.007. Epub 2011 Jan 13. J Mol Biol. 2011. PMID: 21237173 Review.
Cited by
-
Optimizing Polychlorinated Biphenyl Degradation by Flavonoid-Induced Cells of the Rhizobacterium Rhodococcus erythropolis U23A.PLoS One. 2015 May 13;10(5):e0126033. doi: 10.1371/journal.pone.0126033. eCollection 2015. PLoS One. 2015. PMID: 25970559 Free PMC article.
-
Structural evidence for substrate-induced synergism and half-sites reactivity in biotin carboxylase.Protein Sci. 2008 Oct;17(10):1706-18. doi: 10.1110/ps.035584.108. Epub 2008 Aug 25. Protein Sci. 2008. PMID: 18725455 Free PMC article.
-
The unusual convergence of steroid catabolic pathways in Mycobacterium abscessus.Proc Natl Acad Sci U S A. 2022 Oct 4;119(40):e2207505119. doi: 10.1073/pnas.2207505119. Epub 2022 Sep 26. Proc Natl Acad Sci U S A. 2022. PMID: 36161908 Free PMC article.
-
The acid-base-nucleophile catalytic triad in ABH-fold enzymes is coordinated by a set of structural elements.PLoS One. 2020 Feb 21;15(2):e0229376. doi: 10.1371/journal.pone.0229376. eCollection 2020. PLoS One. 2020. PMID: 32084230 Free PMC article.
-
Distinctive structural motifs co-ordinate the catalytic nucleophile and the residues of the oxyanion hole in the alpha/beta-hydrolase fold enzymes.Protein Sci. 2019 Feb;28(2):344-364. doi: 10.1002/pro.3527. Epub 2018 Nov 12. Protein Sci. 2019. PMID: 30311984 Free PMC article.
References
-
- Dagley S. Biochemical approach to some problems of environmental pollution. Essays Biochem. 1975;11:81–138. - PubMed
-
- Pieper DH. Aerobic degradation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 2005;67:170–191. - PubMed
-
- Seah SYK, Labbé G, Nerdinger S, Johnson MR, Snieckus V, Eltis LD. Identification of a serine hydrolase as a key determinant in the microbial degradation of polychlorinated biphenyls. J. Biol. Chem. 2000;275:15701–15708. - PubMed
-
- Ohtsubo Y, Kudo T, Tsuda M, Nagata Y. Strategies for bioremediation of polychlorinated biphenyls. Appl. Microbiol. Biotechnol. 2004;65:250–258. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

