Discovery and validation of breast cancer subtypes
- PMID: 16965636
- PMCID: PMC1574316
- DOI: 10.1186/1471-2164-7-231
Discovery and validation of breast cancer subtypes
Erratum in
- BMC Genomics. 2007 Apr 13;8(1):101
Abstract
Background: Previous studies demonstrated breast cancer tumor tissue samples could be classified into different subtypes based upon DNA microarray profiles. The most recent study presented evidence for the existence of five different subtypes: normal breast-like, basal, luminal A, luminal B, and ERBB2+.
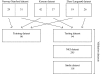
Results: Based upon the analysis of 599 microarrays (five separate cDNA microarray datasets) using a novel approach, we present evidence in support of the most consistently identifiable subtypes of breast cancer tumor tissue microarrays being: ESR1+/ERBB2-, ESR1-/ERBB2-, and ERBB2+ (collectively called the ESR1/ERBB2 subtypes). We validate all three subtypes statistically and show the subtype to which a sample belongs is a significant predictor of overall survival and distant-metastasis free probability.
Conclusion: As a consequence of the statistical validation procedure we have a set of centroids which can be applied to any microarray (indexed by UniGene Cluster ID) to classify it to one of the ESR1/ERBB2 subtypes. Moreover, the method used to define the ESR1/ERBB2 subtypes is not specific to the disease. The method can be used to identify subtypes in any disease for which there are at least two independent microarray datasets of disease samples.
Figures



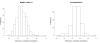

References
-
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge Ø, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. - DOI - PubMed
-
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
-
- Sørlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lønning PE, Brown PO, Børresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci U S A. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. - DOI - PMC - PubMed
-
- Kapp AV, Tibshirani R. Are clusters found in one dataset present in another dataset? Biostatistics. 2006 - PubMed
-
- Fletcher G, Patel S, Tyson K, Adam P, Schenker M, Loader J, Daviet L, Legrain P, Parekh R, Harris A, Terrett J. hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumors and interact with metastasis gene C4.4a and dystroglycan. Br J Cancer. 2003;88:579–585. doi: 10.1038/sj.bjc.6600740. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

