Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes
- PMID: 16966375
- PMCID: PMC1636773
- DOI: 10.1128/MCB.00671-06
Requirement of hCenexin for proper mitotic functions of polo-like kinase 1 at the centrosomes
Abstract
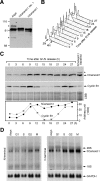
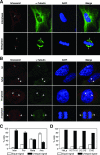
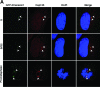
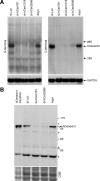
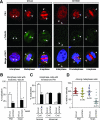
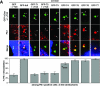
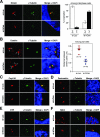
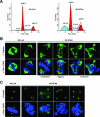
Outer dense fiber 2 (Odf2) was initially identified as a major component of sperm tail cytoskeleton and later was suggested to be a widespread component of centrosomal scaffold that preferentially associates with the appendages of the mother centrioles in somatic cells. Here we report the identification of two Odf2-related centrosomal components, hCenexin1 and hCenexin1 variant 1, that possess a unique C-terminal extension. Our results showed that hCenexin1 is the major isoform expressed in HeLa cells, whereas hOdf2 is not detectably expressed. Mammalian polo-like kinase 1 (Plk1) is critical for proper mitotic progression, and its association with the centrosome is important for microtubule nucleation and function. Interestingly, depletion of hCenexin1 by RNA interference (RNAi) delocalized Plk1 from the centrosomes and the C-terminal extension of hCenexin1 was crucial to recruit Plk1 to the centrosomes through a direct interaction with the polo-box domain of Plk1. Consistent with these findings, the hCenexin1 RNAi cells exhibited weakened gamma-tubulin localization and chromosome segregation defects. We propose that hCenexin1 is a critical centrosomal component whose C-terminal extension is required for proper recruitment of Plk1 and other components crucial for normal mitosis. Our results further suggest that the anti-Odf2 immunoreactive centrosomal antigen previously detected in non-germ line cells is likely hCenexin1.
Figures






References
-
- Arnaud, L., J. Pines, and E. A. Nigg. 1998. GFP tagging reveals human Polo-like kinase 1 at the kinetochore/centromere region of mitotic chromosomes. Chromosoma 107:424-429. - PubMed
-
- Barr, F. A., H. H. Sillje, and E. A. Nigg. 2004. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5:429-440. - PubMed
-
- Brohmann, H., S. Pinnecke, and S. Hoyer-Fender. 1997. Identification and characterization of new cDNAs encoding outer dense fiber proteins of rat sperm. J. Biol. Chem. 272:10327-10332. - PubMed
-
- Casenghi, M., P. Meraldi, U. Weinhart, P. I. Duncan, R. Korner, and E. A. Nigg. 2003. Polo-like kinase 1 regulates Nlp, a centrosome protein involved in microtubule nucleation. Dev. Cell 5:113-125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
