The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation
- PMID: 16966420
- PMCID: PMC2064333
- DOI: 10.1083/jcb.200603042
The centromere-specific histone variant Cse4p (CENP-A) is essential for functional chromatin architecture at the yeast 2-microm circle partitioning locus and promotes equal plasmid segregation
Abstract
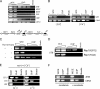
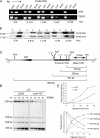
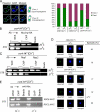
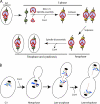
The centromere protein A homologue Cse4p is required for kinetochore assembly and faithful chromosome segregation in Saccharomyces cerevisiae. It has been regarded as the exquisite hallmark of centromeric chromatin. We demonstrate that Cse4 resides at the partitioning locus STB of the 2-microm plasmid. Cse4p-STB association is absolutely dependent on the plasmid partitioning proteins Rep1p and Rep2p and the integrity of the mitotic spindle. The kinetochore mutation ndc10-1 excludes Cse4p from centromeres without dislodging it from STB. Cse4p-STB association lasts from G1/S through late telophase during the cell cycle. The release of Cse4p from STB chromatin is likely mediated through spindle disassembly. A lack of functional Cse4p disrupts the remodeling of STB chromatin by the RSC2 complex, negates Rep2p binding and cohesin assembly at STB, and causes plasmid missegregation. Poaching of a specific histone variant by the plasmid to mark its partitioning locus with a centromere tag reveals yet another one of the molecular trickeries it performs for achieving chromosome- like fidelity in segregation.
Figures



Comment in
-
A hitchhiker's guide to survival finally makes CENs.J Cell Biol. 2006 Sep 11;174(6):747-9. doi: 10.1083/jcb.200608107. J Cell Biol. 2006. PMID: 16966417 Free PMC article. Review.
Similar articles
-
Mutations in a partitioning protein and altered chromatin structure at the partitioning locus prevent cohesin recruitment by the Saccharomyces cerevisiae plasmid and cause plasmid missegregation.Mol Cell Biol. 2004 Jun;24(12):5290-303. doi: 10.1128/MCB.24.12.5290-5303.2004. Mol Cell Biol. 2004. PMID: 15169893 Free PMC article.
-
A novel role for the mitotic spindle during DNA segregation in yeast: promoting 2 microm plasmid-cohesin association.Mol Cell Biol. 2005 May;25(10):4283-98. doi: 10.1128/MCB.25.10.4283-4298.2005. Mol Cell Biol. 2005. PMID: 15870297 Free PMC article.
-
Cse4 (CenH3) association with the Saccharomyces cerevisiae plasmid partitioning locus in its native and chromosomally integrated states: implications in centromere evolution.Mol Cell Biol. 2011 Mar;31(5):1030-40. doi: 10.1128/MCB.01191-10. Epub 2010 Dec 20. Mol Cell Biol. 2011. PMID: 21173161 Free PMC article.
-
Protein kinases in mitotic phosphorylation of budding yeast CENP-A.Curr Genet. 2019 Dec;65(6):1325-1332. doi: 10.1007/s00294-019-00997-5. Epub 2019 May 22. Curr Genet. 2019. PMID: 31119371 Review.
-
Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae.Biochim Biophys Acta. 2012 Jul;1819(7):776-83. doi: 10.1016/j.bbagrm.2012.02.008. Epub 2012 Feb 16. Biochim Biophys Acta. 2012. PMID: 22366340 Free PMC article. Review.
Cited by
-
Centromere identity: a challenge to be faced.Mol Genet Genomics. 2010 Aug;284(2):75-94. doi: 10.1007/s00438-010-0553-4. Epub 2010 Jun 29. Mol Genet Genomics. 2010. PMID: 20585957 Review.
-
Parasitic plasmids are anchored to inactive regions of eukaryotic chromosomes through a nucleosome signal.EMBO J. 2025 Apr;44(7):2134-2156. doi: 10.1038/s44318-025-00389-1. Epub 2025 Feb 27. EMBO J. 2025. PMID: 40016420 Free PMC article.
-
The partitioning and copy number control systems of the selfish yeast plasmid: an optimized molecular design for stable persistence in host cells.Microbiol Spectr. 2014 Oct;2(5):10.1128/microbiolspec.PLAS-0003-2013. doi: 10.1128/microbiolspec.PLAS-0003-2013. Microbiol Spectr. 2014. PMID: 25541598 Free PMC article. Review.
-
A selfish DNA element engages a meiosis-specific motor and telomeres for germ-line propagation.J Cell Biol. 2014 Jun 9;205(5):643-61. doi: 10.1083/jcb.201312002. J Cell Biol. 2014. PMID: 24914236 Free PMC article.
-
Endogenous transcription at the centromere facilitates centromere activity in budding yeast.Curr Biol. 2011 Oct 25;21(20):1695-703. doi: 10.1016/j.cub.2011.08.056. Epub 2011 Oct 13. Curr Biol. 2011. PMID: 22000103 Free PMC article.
References
-
- Bouck, D., and K. Bloom. 2005. The role of centromere-binding factor 3 (CBF3) in spindle stability, cytokinesis, and kinetochore attachment. Biochem. Cell Biol. 83:696–702. - PubMed
-
- Broach, J.R., J.F. Atkins, C. McGill, and L. Chow. 1979. Identification and mapping of the transcriptional and translational products of the yeast plasmid, 2 micron circle. Cell. 16:827–839. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

