Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors
- PMID: 16968137
- PMCID: PMC1563492
- DOI: 10.1371/journal.pbio.0040339
Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors
Abstract
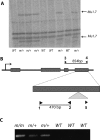
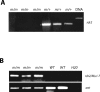
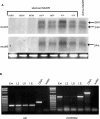
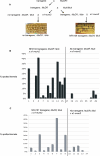
Paramutation and transposon silencing are two epigenetic phenomena that have intrigued and puzzled geneticists for decades. Each involves heritable changes in gene activity without changes in DNA sequence. Here we report the cloning of a gene whose activity is required for the maintenance of both silenced transposons and paramutated color genes in maize. We show that this gene, Mop1 (Mediator of paramutation1) codes for a putative RNA-dependent RNA polymerase, whose activity is required for the production of small RNAs that correspond to the MuDR transposon sequence. We also demonstrate that although Mop1 is required to maintain MuDR methylation and silencing, it is not required for the initiation of heritable silencing. In contrast, we present evidence that a reduction in the transcript level of a maize homolog of the nucleosome assembly protein 1 histone chaperone can reduce the heritability of MuDR silencing. Together, these data suggest that the establishment and maintenance of MuDR silencing have distinct requirements.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



Comment in
-
Transposon silencing keeps jumping genes in their place.PLoS Biol. 2006 Oct;4(10):e353. doi: 10.1371/journal.pbio.0040353. Epub 2006 Sep 12. PLoS Biol. 2006. PMID: 20076477 Free PMC article. No abstract available.
Similar articles
-
Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication.Nat Genet. 2005 Jun;37(6):641-4. doi: 10.1038/ng1576. Epub 2005 May 22. Nat Genet. 2005. PMID: 15908951
-
RNA-directed DNA methylation prevents rapid and heritable reversal of transposon silencing under heat stress in Zea mays.PLoS Genet. 2021 Jun 14;17(6):e1009326. doi: 10.1371/journal.pgen.1009326. eCollection 2021 Jun. PLoS Genet. 2021. PMID: 34125827 Free PMC article.
-
Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays.Genetics. 2003 Oct;165(2):781-97. doi: 10.1093/genetics/165.2.781. Genetics. 2003. PMID: 14573488 Free PMC article.
-
Paramutation: a trans-homolog interaction affecting heritable gene regulation.Curr Opin Plant Biol. 2012 Nov;15(5):536-43. doi: 10.1016/j.pbi.2012.09.003. Epub 2012 Sep 24. Curr Opin Plant Biol. 2012. PMID: 23017240 Review.
-
Paramutation in maize: RNA mediated trans-generational gene silencing.Curr Opin Genet Dev. 2010 Apr;20(2):156-63. doi: 10.1016/j.gde.2010.01.008. Epub 2010 Feb 12. Curr Opin Genet Dev. 2010. PMID: 20153628 Free PMC article. Review.
Cited by
-
Epigenetic reprogramming during vegetative phase change in maize.Proc Natl Acad Sci U S A. 2010 Dec 21;107(51):22184-9. doi: 10.1073/pnas.1016884108. Epub 2010 Dec 6. Proc Natl Acad Sci U S A. 2010. PMID: 21135217 Free PMC article.
-
The Role of Transposable Elements in Speciation.Genes (Basel). 2018 May 15;9(5):254. doi: 10.3390/genes9050254. Genes (Basel). 2018. PMID: 29762547 Free PMC article. Review.
-
Three groups of transposable elements with contrasting copy number dynamics and host responses in the maize (Zea mays ssp. mays) genome.PLoS Genet. 2014 Apr 17;10(4):e1004298. doi: 10.1371/journal.pgen.1004298. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24743518 Free PMC article.
-
Maize RNA polymerase IV defines trans-generational epigenetic variation.Plant Cell. 2013 Mar;25(3):808-19. doi: 10.1105/tpc.112.107680. Epub 2013 Mar 19. Plant Cell. 2013. PMID: 23512852 Free PMC article.
-
A comparative approach to the principal mechanisms of different memory systems.Naturwissenschaften. 2009 Dec;96(12):1373-84. doi: 10.1007/s00114-009-0591-0. Epub 2009 Aug 13. Naturwissenschaften. 2009. PMID: 19680619 Review.
References
-
- Ringrose L, Paro R. Remembering silence. Bioessays. 2001;23:566–570. - PubMed
-
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. - PubMed
-
- Brink RA, Styles ED, Axtell JD. Paramutation: Directed genetic change. Paramutation occurs in somatic cells and heritably alters the functional state of a locus. Science. 1968;159:161–170. - PubMed
-
- Chandler VL, Eggleston WB, Dorweiler JE. Paramutation in maize. Plant Mol Biol. 2000;43:121–145. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

