Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase
- PMID: 16968695
- PMCID: PMC1769310
- DOI: 10.1074/jbc.M606654200
Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase
Abstract
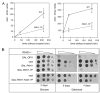
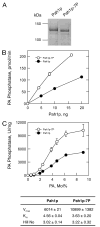
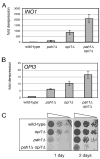
Phosphorylation of the conserved lipin Pah1p/Smp2p in Saccharomyces cerevisiae was previously shown to control transcription of phospholipid biosynthetic genes and nuclear structure by regulating the amount of membrane present at the nuclear envelope (Santos-Rosa, H., Leung, J., Grimsey, N., Peak-Chew, S., and Siniossoglou, S. (2005) EMBO J. 24, 1931-1941). A recent report identified Pah1p as a Mg2+-dependent phosphatidate (PA) phosphatase that regulates de novo lipid synthesis (Han G.-S., Wu, W. I., and Carman, G. M. (2006) J. Biol. Chem. 281, 9210-9218). In this work we use a combination of mass spectrometry and systematic mutagenesis to identify seven Ser/Thr-Pro motifs within Pah1p that are phosphorylated in vivo. We show that phosphorylation on these sites is required for the efficient transcriptional derepression of key enzymes involved in phospholipid biosynthesis. The phosphorylation-deficient Pah1p exhibits higher PA phosphatase-specific activity than the wild-type Pah1p, indicating that phosphorylation of Pah1p controls PA production. Opi1p is a transcriptional repressor of phospholipid biosynthetic genes, responding to PA levels. Genetic analysis suggests that Pah1p regulates transcription of these genes through both Opi1p-dependent and -independent mechanisms. We also provide evidence that derepression of phospholipid biosynthetic genes is not sufficient to induce the nuclear membrane expansion shown in the pah1delta cells.
Figures




Similar articles
-
Pho85p-Pho80p phosphorylation of yeast Pah1p phosphatidate phosphatase regulates its activity, location, abundance, and function in lipid metabolism.J Biol Chem. 2012 Mar 30;287(14):11290-301. doi: 10.1074/jbc.M112.346023. Epub 2012 Feb 9. J Biol Chem. 2012. PMID: 22334681 Free PMC article.
-
Host Pah1p phosphatidate phosphatase limits viral replication by regulating phospholipid synthesis.PLoS Pathog. 2018 Apr 12;14(4):e1006988. doi: 10.1371/journal.ppat.1006988. eCollection 2018 Apr. PLoS Pathog. 2018. PMID: 29649282 Free PMC article.
-
Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of SER(602), THR(723), AND SER(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase.J Biol Chem. 2011 Jan 14;286(2):1486-98. doi: 10.1074/jbc.M110.155598. Epub 2010 Nov 16. J Biol Chem. 2011. PMID: 21081492 Free PMC article.
-
Phosphatidate phosphatase, a key regulator of lipid homeostasis.Biochim Biophys Acta. 2013 Mar;1831(3):514-22. doi: 10.1016/j.bbalip.2012.08.006. Epub 2012 Aug 14. Biochim Biophys Acta. 2013. PMID: 22910056 Free PMC article. Review.
-
Lipins, lipids and nuclear envelope structure.Traffic. 2009 Sep;10(9):1181-7. doi: 10.1111/j.1600-0854.2009.00923.x. Epub 2009 May 20. Traffic. 2009. PMID: 19490535 Review.
Cited by
-
Uncovering the Role of the Yeast Lysine Acetyltransferase NuA4 in the Regulation of Nuclear Shape and Lipid Metabolism.Mol Cell Biol. 2024;44(7):273-288. doi: 10.1080/10985549.2024.2366206. Epub 2024 Jul 4. Mol Cell Biol. 2024. PMID: 38961766 Free PMC article.
-
Lipid droplet dynamics in budding yeast.Cell Mol Life Sci. 2015 Jul;72(14):2677-95. doi: 10.1007/s00018-015-1903-5. Epub 2015 Apr 18. Cell Mol Life Sci. 2015. PMID: 25894691 Free PMC article. Review.
-
Fat-regulating phosphatidic acid phosphatase: a review of its roles and regulation in lipid homeostasis.J Lipid Res. 2019 Jan;60(1):2-6. doi: 10.1194/jlr.S087452. Epub 2018 Dec 7. J Lipid Res. 2019. PMID: 30530634 Free PMC article. Review.
-
Protein kinase A phosphorylates the Nem1-Spo7 protein phosphatase complex that regulates the phosphorylation state of the phosphatidate phosphatase Pah1 in yeast.J Biol Chem. 2018 Oct 12;293(41):15801-15814. doi: 10.1074/jbc.RA118.005348. Epub 2018 Sep 10. J Biol Chem. 2018. PMID: 30201607 Free PMC article.
-
Regulation of phospholipid synthesis in yeast.J Lipid Res. 2009 Apr;50 Suppl(Suppl):S69-73. doi: 10.1194/jlr.R800043-JLR200. Epub 2008 Oct 27. J Lipid Res. 2009. PMID: 18955729 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

