The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor
- PMID: 16968770
- PMCID: PMC1599896
- DOI: 10.1073/pnas.0606384103
The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor
Abstract
Translational initiation factor 2 (IF2) is a guanine nucleotide-binding protein that can bind guanosine 3',5'-(bis) diphosphate (ppGpp), an alarmone involved in stringent response in bacteria. In cells growing under optimal conditions, the GTP concentration is very high, and that of ppGpp very low. However, under stress conditions, the GTP concentration may decline by as much as 50%, and that of ppGpp can attain levels comparable to those of GTP. Here we show that IF2 binds ppGpp at the same nucleotide-binding site and with similar affinity as GTP. Thus, GTP and the alarmone ppGpp can be considered two alternative physiologically relevant IF2 ligands. ppGpp interferes with IF2-dependent initiation complex formation, severely inhibits initiation dipeptide formation, and blocks the initiation step of translation. Our data suggest that IF2 has the properties of a cellular metabolic sensor and regulator that oscillates between an active GTP-bound form under conditions allowing active protein syntheses and an inactive ppGpp-bound form when shortage of nutrients would be detrimental, if not accompanied by slackening of this synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

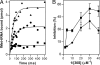



 ▴), association of 50S subunit and formation of 70SIC, GTP (▴) hydrolysis to GDP (
▴), association of 50S subunit and formation of 70SIC, GTP (▴) hydrolysis to GDP ( ), IF2 dissociation (
), IF2 dissociation ( ), and initiation dipeptide formation. Amino acid starvation during elongation triggers RelA-dependent synthesis of ppGpp (■), which inhibits stable RNA transcription (Lower) and, as shown in this study, the initiation functions of IF2 (30SIC formation and initiation dipeptide formation). A possible function of translation initiation intermediates containing IF2-ppGpp (
), and initiation dipeptide formation. Amino acid starvation during elongation triggers RelA-dependent synthesis of ppGpp (■), which inhibits stable RNA transcription (Lower) and, as shown in this study, the initiation functions of IF2 (30SIC formation and initiation dipeptide formation). A possible function of translation initiation intermediates containing IF2-ppGpp ( ) in feedback inhibition of stable RNA transcription is suggested by earlier reports that components of the translation initiation apparatus play an active part in this regulation. In particular, IF2, fMet-tRNA, and ppGpp were found to interact with the RNA polymerase and influence its activity at stable RNA promoters (38, 39), whereas IF2 (40), IF3 (41), and initiation-competent 30S subunits (42) were shown to be required for feedback repression of stable RNA transcription.
) in feedback inhibition of stable RNA transcription is suggested by earlier reports that components of the translation initiation apparatus play an active part in this regulation. In particular, IF2, fMet-tRNA, and ppGpp were found to interact with the RNA polymerase and influence its activity at stable RNA promoters (38, 39), whereas IF2 (40), IF3 (41), and initiation-competent 30S subunits (42) were shown to be required for feedback repression of stable RNA transcription.References
-
- Gualerzi CO, Brandi L, Caserta E, La Teana A, Spurio R, Tomšic J, Pon CL. In: The Ribosome: Structure, Function, Antibiotics, and Cellular Interactions. Garrett RA, Douthwaite SR, Liljas A, Matheson AT, Moore PB, Noller HF, editors. Washington, DC: ASM Press; 2000. pp. 477–494.
-
- Gualerzi CO, Brandi L, Caserta E, Garofalo C, Lammi M, La Teana A, Petrelli D, Spurio R, Tomšic J, Pon CL. Cold Spring Harbor Symp Quant Biol. 2001;66:363–376. - PubMed
-
- Boelens R, Gualerzi CO. Curr Protein Pept Sci. 2002;3:107–119. - PubMed
-
- Ramakrishnan V. Cell. 2002;108:557–572. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

