MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line
- PMID: 16968818
- PMCID: PMC2435371
- DOI: 10.1242/dev.02584
MES-4: an autosome-associated histone methyltransferase that participates in silencing the X chromosomes in the C. elegans germ line
Abstract
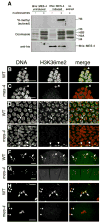
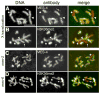
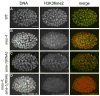
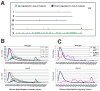
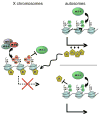
Germ cell development in C. elegans requires that the X chromosomes be globally silenced during mitosis and early meiosis. We previously found that the nuclear proteins MES-2, MES-3, MES-4 and MES-6 regulate the different chromatin states of autosomes versus X chromosomes and are required for germline viability. Strikingly, the SET-domain protein MES-4 is concentrated on autosomes and excluded from the X chromosomes. Here, we show that MES-4 has histone H3 methyltransferase (HMT) activity in vitro, and is required for histone H3K36 dimethylation in mitotic and early meiotic germline nuclei and early embryos. MES-4 appears unlinked to transcription elongation, thus distinguishing it from other known H3K36 HMTs. Based on microarray analysis, loss of MES-4 leads to derepression of X-linked genes in the germ line. We discuss how an autosomally associated HMT may participate in silencing genes on the X chromosome, in coordination with the direct silencing effects of the other MES proteins.
Figures



References
-
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem. 2005;280:17732–17736. - PubMed
-
- Baugh LR, Hill AA, Slonim DK, Brown EL, Hunter CP. Composition and dynamics of the Caenorhabditis elegans early embryonic transcriptome. Development. 2003;130:889–900. - PubMed
-
- Bender LB, Cao R, Zhang Y, Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

