Principles of microRNA regulation of a human cellular signaling network
- PMID: 16969338
- PMCID: PMC1681519
- DOI: 10.1038/msb4100089
Principles of microRNA regulation of a human cellular signaling network
Abstract
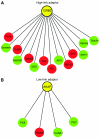
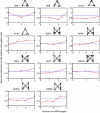
MicroRNAs (miRNAs) are endogenous approximately 22-nucleotide RNAs, which suppress gene expression by selectively binding to the 3'-noncoding region of specific messenger RNAs through base-pairing. Given the diversity and abundance of miRNA targets, miRNAs appear to functionally interact with various components of many cellular networks. By analyzing the interactions between miRNAs and a human cellular signaling network, we found that miRNAs predominantly target positive regulatory motifs, highly connected scaffolds and most downstream network components such as signaling transcription factors, but less frequently target negative regulatory motifs, common components of basic cellular machines and most upstream network components such as ligands. In addition, when an adaptor has potential to recruit more downstream components, these components are more frequently targeted by miRNAs. This work uncovers the principles of miRNA regulation of signal transduction networks and implies a potential function of miRNAs for facilitating robust transitions of cellular response to extracellular signals and maintaining cellular homeostasis.
Figures

References
-
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355 - PubMed
-
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA (2004) Structure and evolution of transcriptional regulatory networks. Curr Opin Struct Biol 14: 283–291 - PubMed
-
- Barabasi AL, Oltvai ZN (2004) Network biology: understanding the cell's functional organization. Nat Rev Genet 5: 101–113 - PubMed
-
- Dekel E, Mangan S, Alon U (2005) Environmental selection of the feed-forward loop circuit in gene-regulation networks. Phys Biol 2: 81–88 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

