Characterization of the aldo-keto reductase 1C gene cluster on pig chromosome 10: possible associations with reproductive traits
- PMID: 16970816
- PMCID: PMC1586007
- DOI: 10.1186/1746-6148-2-28
Characterization of the aldo-keto reductase 1C gene cluster on pig chromosome 10: possible associations with reproductive traits
Abstract
Background: The rate of pubertal development and weaning to estrus interval are correlated and affect reproductive efficiency of swine. Quantitative trait loci (QTL) for age of puberty, nipple number and ovulation rate have been identified in Meishan crosses on pig chromosome 10q (SSC10) near the telomere, which is homologous to human chromosome 10p15 and contains an aldo-keto reductase (AKR) gene cluster with at least six family members. AKRs are tissue-specific hydroxysteroid dehydrogenases that interconvert weak steroid hormones to their more potent counterparts and regulate processes involved in development, homeostasis and reproduction. Because of their location in the swine genome and their implication in reproductive physiology, this gene cluster was characterized and evaluated for effects on reproductive traits in swine.
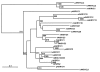
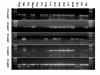
Results: Screening the porcine CHORI-242 BAC library with a full-length AKR1C4 cDNA identified 7 positive clones and sample sequencing of 5 BAC clones revealed 5 distinct AKR1C genes (AKR1CL2 and AKR1C1 through 4), which mapped to 126-128 cM on SSC10. Using the IMpRH7000rad and IMNpRH212000rad radiation hybrid panels, these 5 genes mapped between microsatellite markers SWR67 and SW2067. Comparison of sequence data with the porcine BAC fingerprint map show that the cluster of genes resides in a 300 kb region. Twelve SNPs were genotyped in gilts observed for age at first estrus and ovulation rate from the F8 and F10 generations of one-quarter Meishan descendants of the USMARC resource population. Age at puberty, nipple number and ovulation rate data were analyzed for association with genotypes by MTDFREML using an animal model. One SNP, a phenylalanine to isoleucine substitution in AKR1C2, was associated with age of puberty (p = 0.07) and possibly ovulation rate (p = 0.102). Two SNP in AKR1C4 were significantly associated with nipple number (p </= 0.03) and another possibly associated with age at puberty (p = 0.09).
Conclusion: AKR1C genotypes were associated with nipple number as well as possible effects on age at puberty and ovulation rate. The estimated effects of AKR1C genotypes on these traits suggest that the SNPs are in incomplete linkage disequilibrium with the causal mutations that affect reproductive traits in swine. Further investigations are necessary to identify these mutations and understand how these AKR1C genes affect these important reproductive traits. The nucleotide sequence data reported have been submitted to GenBank and assigned accession numbers [GenBank:DQ474064-DQ474068, GenBank:DQ494488-DQ494490 and GenBank:DQ487182-DQ487184].
Figures



References
-
- Rauw WM, Kanis E, Noordhuizen-Stassen EN, Grommers FJ. Undesirable side effects of selection for high production efficiency in farm animals: a review. Livestock Production Science. 1998;56:15–33. doi: 10.1016/S0301-6226(98)00147-X. - DOI
-
- Sterning M, Rydhmer L, Eliasson-Selling L. Relationships between age at puberty and interval from weaning to estrus and between estrus signs at puberty and after the first weaning in pigs. Journal of Animal Science. 1998;76:353–359. - PubMed
-
- Cassady JP, Johnson RK, Pomp D, Rohrer GA, Van Vleck LD, Spiegel EK, Gilson KM. Identification of quantitative trait loci affecting reproduction in pigs. Journal of Animal Science. 2001;79:623–633. - PubMed
-
- Rohrer GA, Ford JJ, Wise TH, Vallet JL, Christenson RK. Identification of quantitative trait loci affecting female reproductive traits in a multigeneration Meishan-White composite swine population. Journal of Animal Science. 1999;77:1385–1391. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

