A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites
- PMID: 16971446
- PMCID: PMC1642152
- DOI: 10.1128/JVI.01737-06
A high-throughput method for cloning and sequencing human immunodeficiency virus type 1 integration sites
Abstract
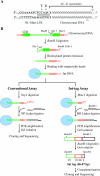
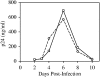
Integration of retroviral DNA is nonspecific and can occur at many sites throughout chromosomes. However, the process is not uniformly distributed, and both hot and cold spots for integration exist. The mechanism that determines target site specificity is not well understood. Because of the nonspecific and widespread nature of integration, studies analyzing the mechanism and factors that control target site selection require the collection and analysis of a large library of human immunodeficiency virus type 1 (HIV-1) proviral clones. Such analyses are time-consuming and labor-intensive using conventional means. We have developed an efficient and high-throughput method of sequencing and mapping a large number of independent integration sites in the absence of any selection or bias. The new assay involves the use of a modified HIV-1 (NL-Mme) containing a type IIS restriction site, MmeI, at the right end of viral DNA. Digestion of genomic DNA from NL-Mme-infected cells generated viral DNA-containing fragments of a discrete size. Subsequent ligation-mediated PCR yielded short integration site fragments termed Int-tags, which were concatemerized for determining multiple integration sites in a single sequencing reaction. Analysis of chromosomal features and sequence preference associated with integration events confirmed the validity of the new high-throughput assay. The assay will aid the effort in understanding the mechanisms of target site selection during HIV-1 DNA integration, and the described methodology can be adapted easily to integration site studies involving other retroviruses and transposons.
Figures
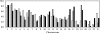
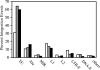
Similar articles
-
Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells.J Virol. 2006 Feb;80(4):1939-48. doi: 10.1128/JVI.80.4.1939-1948.2006. J Virol. 2006. PMID: 16439549 Free PMC article.
-
Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription.J Virol. 2005 Jun;79(11):6610-9. doi: 10.1128/JVI.79.11.6610-6619.2005. J Virol. 2005. PMID: 15890899 Free PMC article.
-
Identification of HIV integration sites in infected host genomic DNA.Methods. 2011 Jan;53(1):39-46. doi: 10.1016/j.ymeth.2010.04.004. Epub 2010 Apr 10. Methods. 2011. PMID: 20385239
-
Sites of retroviral DNA integration: From basic research to clinical applications.Crit Rev Biochem Mol Biol. 2016;51(1):26-42. doi: 10.3109/10409238.2015.1102859. Epub 2015 Oct 28. Crit Rev Biochem Mol Biol. 2016. PMID: 26508664 Free PMC article. Review.
-
HIV-1 gene expression: lessons from provirus and non-integrated DNA.Retrovirology. 2004 Jun 25;1:13. doi: 10.1186/1742-4690-1-13. Retrovirology. 2004. PMID: 15219234 Free PMC article. Review.
Cited by
-
Retroviral integration site selection.Viruses. 2010 Jan;2(1):111-130. doi: 10.3390/v2010111. Epub 2010 Jan 12. Viruses. 2010. PMID: 21994603 Free PMC article.
-
Fidelity of target site duplication and sequence preference during integration of xenotropic murine leukemia virus-related virus.PLoS One. 2010 Apr 20;5(4):e10255. doi: 10.1371/journal.pone.0010255. PLoS One. 2010. PMID: 20421928 Free PMC article.
-
Specific insertions of zinc finger domains into Gag-Pol yield engineered retroviral vectors with selective integration properties.Proc Natl Acad Sci U S A. 2010 Jul 13;107(28):12475-80. doi: 10.1073/pnas.1001402107. Epub 2010 Jun 28. Proc Natl Acad Sci U S A. 2010. PMID: 20616052 Free PMC article.
-
An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors.Proc Natl Acad Sci U S A. 2007 Jan 30;104(5):1655-60. doi: 10.1073/pnas.0610291104. Epub 2007 Jan 18. Proc Natl Acad Sci U S A. 2007. PMID: 17234809 Free PMC article.
-
Natural and engineered nicking endonucleases--from cleavage mechanism to engineering of strand-specificity.Nucleic Acids Res. 2011 Jan;39(1):1-18. doi: 10.1093/nar/gkq742. Epub 2010 Aug 30. Nucleic Acids Res. 2011. PMID: 20805246 Free PMC article. Review.
References
-
- Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1999. Current protocols in molecular biology. Wiley, New York, N.Y.
-
- Brown, P. O. 1997. Integration, p. 161-204. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
-
- Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

