Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH
- PMID: 16971452
- PMCID: PMC1642150
- DOI: 10.1128/JVI.01287-06
Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH
Abstract
Human metapneumovirus (HMPV) is a recently described human pathogen of the pneumovirus subfamily within the paramyxovirus family. HMPV infection is prevalent worldwide and is associated with severe respiratory disease, particularly in infants. The HMPV fusion protein (F) amino acid sequence contains features characteristic of other paramyxovirus F proteins, including a putative cleavage site and potential N-linked glycosylation sites. Propagation of HMPV in cell culture requires exogenous trypsin, which cleaves the F protein, and HMPV, like several other pneumoviruses, is infectious in the absence of its attachment protein (G). However, little is known about HMPV F-promoted fusion, since the HMPV glycoproteins have yet to be analyzed separately from the virus. Using syncytium and luciferase reporter gene fusion assays, we determined the basic requirements for HMPV F protein-promoted fusion in transiently transfected cells. Our data indicate that proteolytic cleavage of the F protein is a stringent requirement for fusion and that the HMPV G protein does not significantly enhance fusion. Unexpectedly, we also found that fusion can be detected only when transfected cells are treated with trypsin and exposed to low pH, indicating that this viral fusion protein may function in a manner unique among the paramyxoviruses. We also analyzed the F protein cleavage site and three potential N-linked glycosylation sites by mutagenesis. Mutations in the cleavage site designed to facilitate endogenous cleavage did so with low efficiency, and our data suggest that all three N-glycosylation sites are utilized and that each affects cleavage and fusion to various degrees.
Figures

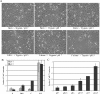


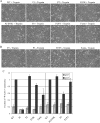
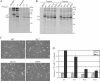
References
-
- Bagai, S., and R. A. Lamb. 1995. Individual roles of N-linked oligosaccharide chains in intracellular transport of the paramyxovirus SV5 fusion protein. Virology 209:250-256. - PubMed
-
- Barretto, N., L. K. Hallak, and M. E. Peeples. 2003. Neuraminidase treatment of respiratory syncytial virus-infected cells or virions, but not target cells, enhances cell-cell fusion and infection. Virology 313:33-43. - PubMed
-
- Biacchesi, S., Q. N. Pham, M. H. Skiadopoulos, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2005. Infection of nonhuman primates with recombinant human metapneumovirus lacking the SH, G, or M2-2 protein categorizes each as a nonessential accessory protein and identifies vaccine candidates. J. Virol. 79:12608-12613. - PMC - PubMed
-
- Biacchesi, S., Q. N. Pham, M. H. Skiadopoulos, B. R. Murphy, P. L. Collins, and U. J. Buchholz. 2006. Modification of the trypsin-dependent cleavage activation site of the human metapneumovirus fusion protein to be trypsin independent does not increase replication or spread in rodents or nonhuman primates. J. Virol. 80:5798-5806. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

