Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice
- PMID: 16973545
- PMCID: PMC1617232
- DOI: 10.1128/JVI.00605-06
Cidofovir resistance in vaccinia virus is linked to diminished virulence in mice
Abstract
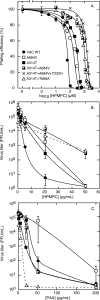
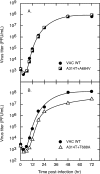
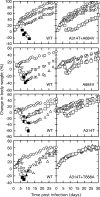
Cidofovir [(S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine (HPMPC)] is recognized as a promising drug for the treatment of poxvirus infections, but drug resistance can arise by a mechanism that is poorly understood. We show here that in vitro selection for high levels of resistance to HPMPC produces viruses encoding two substitution mutations in the virus DNA polymerase (E9L) gene. These mutations are located within the regions of the gene encoding the 3'-5' exonuclease (A314T) and polymerase (A684V) catalytic domains. These mutant viruses exhibited cross-resistance to other nucleoside phosphonate drugs, while they remained sensitive to other unrelated DNA polymerase inhibitors. Marker rescue experiments were used to transfer A314T and/or A684V alleles into a vaccinia virus Western Reserve strain. Either mutation alone could confer a drug resistance phenotype, although the degree of resistance was significantly lower than when virus encoded both mutations. The A684V substitution, but not the A314T change, also conferred a spontaneous mutator phenotype. All of the HPMPC-resistant recombinant viruses exhibited reduced virulence in mice, demonstrating that these E9L mutations are inextricably linked to reduced fitness in vivo. HPMPC, at a dose of 50 mg/kg of body weight/day for 5 days, still protected mice against intranasal challenge with the drug-resistant virus with A314T and A684V mutations. Our studies show that proposed drug therapies offer a reasonable likelihood of controlling orthopoxvirus infections, even if the viruses encode drug resistance markers.
Figures


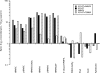



References
-
- Andrei, G., R. Snoeck, E. De Clercq, R. Esnouf, P. Fiten, and G. Opdenakker. 2000. Resistance of herpes simplex virus type 1 against different phosphonylmethoxyalkyl derivatives of purines and pyrimidines due to specific mutations in the viral DNA polymerase gene. J. Gen. Virol. 81:639-648. - PubMed
-
- Blanco, L., A. Bernad, and M. Salas. 1992. Evidence favouring the hypothesis of a conserved 3′-5′ exonuclease active site in DNA-dependent DNA polymerases. Gene 112:139-144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

