Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related L1 protein
- PMID: 16973551
- PMCID: PMC1617236
- DOI: 10.1128/JVI.01149-06
Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related L1 protein
Abstract
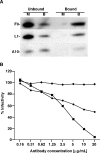
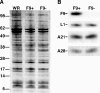
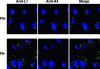
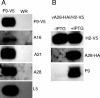
All sequenced poxviruses encode orthologs of the vaccinia virus L1 and F9 proteins, which are structurally similar and share about 20% amino acid identity. We found that F9 further resembles L1 as both proteins are membrane components of the mature virion with similar topologies and induce neutralizing antibodies. In addition, a recombinant vaccinia virus that inducibly expresses F9, like a previously described L1 mutant, had a conditional-lethal phenotype: plaque formation and replication of infectious virus were dependent on added inducer. However, only immature virus particles are made when L1 is repressed, whereas normal-looking intracellular and extracellular virions formed in the absence of F9. Except for the lack of F9, the polypeptide components of such virions were indistinguishable from those of wild-type virus. These F9-deficient virions bound to cells, but their cores did not penetrate into the cytoplasm. Furthermore, cells infected with F9-negative virions did not fuse after a brief low-pH treatment, as did cells infected with virus made in the presence of inducer. In these respects, the phenotype associated with F9 deficiency was identical to that produced by the lack of individual components of a previously described poxvirus entry/fusion complex. Moreover, F9 interacted with proteins of that complex, supporting a related role. Thus, despite the structural relationships of L1 and F9, the two proteins have distinct functions in assembly and entry, respectively.
Figures







References
-
- Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. - PubMed
-
- Aldaz-Carroll, L., J. C. Whitbeck, M. Ponce de Leon, H. Lou, L. K. Pannell, J. Lebowitz, C. Fogg, C. White, B. Moss, G. H. Cohen, and R. J. Eisenberg. 2005. Physical and immunological characterization of a recombinant secreted form of the membrane protein encoded by the vaccinia virus L1R gene. Virology 341:59-71. - PubMed
-
- Armstrong, J. A., D. H. Metz, and M. R. Young. 1973. The mode of entry of vaccinia virus into L cells. J. Gen. Virol. 21:533-537. - PubMed
-
- Broyles, S. S. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293-2303. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

