Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity
- PMID: 16973562
- PMCID: PMC1617272
- DOI: 10.1128/JVI.00141-06
Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity
Abstract
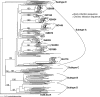
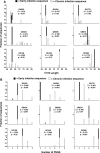
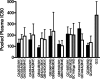
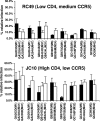
Over the course of infection, human immunodeficiency virus type 1 (HIV-1) continuously adapts to evade the evolving host neutralizing antibody responses. Changes in the envelope variable loop sequences, particularly the extent of glycosylation, have been implicated in antibody escape. To document modifications that potentially influence antibody susceptibility, we compared envelope variable loops 1 and 2 (V1-V2) from multiple sequences isolated at the primary phase of infection to those isolated around 2 to 3 years into the chronic phase of infection in nine women with HIV-1 subtype A. HIV-1 sequences isolated during chronic infection had significantly longer V1-V2 loops, with a significantly higher number of potential N-linked glycosylation sites, than the sequences isolated early in infection. To assess the effects of these V1-V2 changes on antibody neutralization and infectivity, we created chimeric envelope sequences, which incorporated a subject's V1-V2 sequences into a common subtype A envelope backbone and then used them to generate pseudotyped viruses. Compared to the parent virus, the introduction of a subject's early-infection V1-V2 envelope variable loops rendered the chimeric envelope more sensitive to that subject's plasma samples but only to plasma samples collected >6 months after the sequences were isolated. Neutralization was not detected with the same plasma when the early-infection V1-V2 sequences were replaced with chronic-infection V1-V2 sequences, suggesting that changes in V1-V2 contribute to antibody escape. Pseudotyped viruses with V1-V2 segments from different times in infection, however, showed no significant difference in neutralization sensitivity to heterologous pooled plasma, suggesting that viruses with V1-V2 loops from early in infection were not inherently more neutralization sensitive. Pseudotyped viruses bearing chimeric envelopes with early-infection V1-V2 sequences showed a trend in infecting cells with low CD4 concentrations more efficiently, while engineered viruses with V1-V2 sequences isolated during chronic infection were moderately better at infecting cells with low CCR5 concentrations. These studies suggest that changes within the V1-V2 envelope domains over the course of an infection influence sensitivity to autologous neutralizing antibodies and may also impact host receptor/coreceptor interactions.
Figures



References
-
- Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. - PubMed
-
- Blay, W., S. Gnanakaran, B. Foley, N. A. Doria-Rose, B. Korber, and N. Haigwood. 2006. Consistent patterns of change during divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J. Virol. 80:999-1014. - PMC - PubMed
-
- Cheng-Mayer, C., A. Brown, J. Harouse, P. A. Luciw, and A. J. Mayer. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J. Virol. 73:5294-5300. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

