Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor
- PMID: 16977310
- PMCID: PMC1590002
- DOI: 10.1038/sj.emboj.7601334
Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor
Abstract
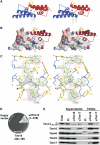
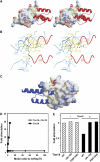
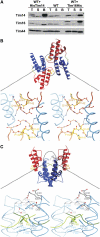
The import motor of the mitochondrial translocase of the inner membrane (TIM23) mediates the ATP-dependent translocation of preproteins into the mitochondrial matrix by cycles of binding to and release from mtHsp70. An essential step of this process is the stimulation of the ATPase activity of mtHsp70 performed by the J cochaperone Tim14. Tim14 forms a complex with the J-like protein Tim16. The crystal structure of this complex shows that the conserved domains of the two proteins have virtually identical folds but completely different surfaces enabling them to perform different functions. The Tim14-Tim16 dimer reveals a previously undescribed arrangement of J and J-like domains. Mutations that destroy the complex between Tim14 and Tim16 are lethal demonstrating that complex formation is an essential requirement for the viability of cells. We further demonstrate tight regulation of the cochaperone activity of Tim14 by Tim16. The first crystal structure of a J domain in complex with a regulatory protein provides new insights into the function of the mitochondrial TIM23 translocase and the Hsp70 chaperone system in general.
Figures



References
-
- Berjanskii M, Riley M, Van Doren SR (2002) Hsc70-interacting HPD loop of the J domain of polyomavirus T antigens fluctuates in ps to ns and micros to ms. J Mol Biol 321: 503–516 - PubMed
-
- Boeke JD, Trueheart J, Natsoulis G, Fink GR (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol 154: 164–175 - PubMed
-
- Bradford MM (1976) A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal Biochem 72: 248–254 - PubMed
-
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 - PubMed
-
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, Pfanner N, Rehling P (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120: 817–829 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

