Opposing roles for Set2 and yFACT in regulating TBP binding at promoters
- PMID: 16977311
- PMCID: PMC1589996
- DOI: 10.1038/sj.emboj.7601333
Opposing roles for Set2 and yFACT in regulating TBP binding at promoters
Abstract
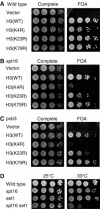
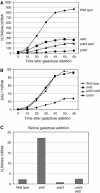
Previous work links histone methylation by Set2 with transcriptional elongation. yFACT (Spt16-Pob3 and Nhp6) reorganizes nucleosomes and functions in both transcriptional initiation and elongation. We show that growth defects caused by spt16 or pob3 mutations can be suppressed by deleting SET2, suggesting that Set2 and yFACT have opposing roles. Set2 methylates K36 of histone H3, and K36 substitutions also suppress yFACT mutations. In contrast, set1 enhances yFACT mutations. Methylation at H3 K4 by Set1 is required for set2 to suppress yFACT defects. We did not detect an elongation defect at an 8 kb ORF in yFACT mutants. Instead, pob3 mutants displayed reduced binding of both pol II and TBP to the GAL1 promoter. Importantly, both GAL1 transcription and promoter binding of pol II and TBP are significantly restored in the pob3 set2 double mutant. Defects caused by an spt16 mutation are enhanced by either TBP or TFIIA mutants. These synthetic defects are suppressed by set2, demonstrating that yFACT and Set2 oppose one another during transcriptional initiation at a step involving DNA binding by TBP and TFIIA.
Figures





References
-
- Ausubel FM, Brent R, Kingston RE, Moore DE, Seidman JG, Smith JA, Struhl K (1987) Current Protocols in Molecular Biology. New York: Wiley and Sons
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

