p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation
- PMID: 16977321
- PMCID: PMC1589997
- DOI: 10.1038/sj.emboj.7601307
p97 functions as an auxiliary factor to facilitate TM domain extraction during CFTR ER-associated degradation
Abstract
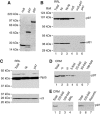
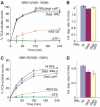
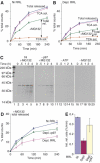
The AAA-ATPase (ATPase associated with various cellular activities) p97 has been implicated in the degradation of misfolded and unassembled proteins in the endoplasmic reticulum (ERAD). To better understand its role in this process, we used a reconstituted cell-free system to define the precise contribution of p97 in degrading immature forms of the polytopic, multi-domain protein CFTR (cystic fibrosis transmembrane conductance regulator). Although p97 augmented both the rate and the extent of CFTR degradation, it was not obligatorily required for ERAD. Only a 50% decrease in degradation was observed in the complete absence of p97. Moreover, p97 specifically stimulated the degradation of CFTR transmembrane (TM) domains but had no effect on isolated cytosolic domains. Consistent with this, p97-mediated extraction of intact TM domains was independent of proteolytic cleavage and influenced by TM segment hydrophobicity, indicating that the relative contribution of p97 is partially determined by substrate stability. Thus, we propose that p97 functions in ERAD as a nonessential but important ancillary component to the proteasome where it facilitates substrate presentation and increases the degradation rate and efficiency of stable (TM) domains.
Figures





References
-
- Babbitt SE, Kiss A, Deffenbaugh AE, Chang YH, Bailly E, Erdjument-Bromage H, Tempst PTB, Sklar LA, Baumler J, Gogol E, Skowyra D (2005) ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell 121: 553–565 - PubMed
-
- Baumeister W, Walz J, Zuehl F, Seemueller E (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell 92: 367–380 - PubMed
-
- Bays N, Hampton R (2002) Cdc48–Ufd1–Npl4: stuck in the middle with Ub. Curr Biol 12: R366–R371 - PubMed
-
- Braun B, Glickman M, Kraft R, Dahlmann B, Kloetzel P-M, Finley D, Schmidt M (1999) The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat Cell Biol 1: 221–226 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

