Transcriptional dynamics of the embryonic stem cell switch
- PMID: 16978048
- PMCID: PMC1570179
- DOI: 10.1371/journal.pcbi.0020123
Transcriptional dynamics of the embryonic stem cell switch
Abstract
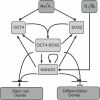
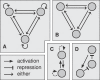
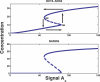
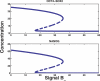
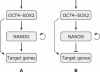
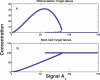
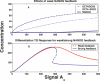
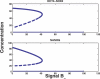
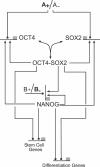
Recent ChIP experiments of human and mouse embryonic stem cells have elucidated the architecture of the transcriptional regulatory circuitry responsible for cell determination, which involves the transcription factors OCT4, SOX2, and NANOG. In addition to regulating each other through feedback loops, these genes also regulate downstream target genes involved in the maintenance and differentiation of embryonic stem cells. A search for the OCT4-SOX2-NANOG network motif in other species reveals that it is unique to mammals. With a kinetic modeling approach, we ascribe function to the observed OCT4-SOX2-NANOG network by making plausible assumptions about the interactions between the transcription factors at the gene promoter binding sites and RNA polymerase (RNAP), at each of the three genes as well as at the target genes. We identify a bistable switch in the network, which arises due to several positive feedback loops, and is switched on/off by input environmental signals. The switch stabilizes the expression levels of the three genes, and through their regulatory roles on the downstream target genes, leads to a binary decision: when OCT4, SOX2, and NANOG are expressed and the switch is on, the self-renewal genes are on and the differentiation genes are off. The opposite holds when the switch is off. The model is extremely robust to parameter changes. In addition to providing a self-consistent picture of the transcriptional circuit, the model generates several predictions. Increasing the binding strength of NANOG to OCT4 and SOX2, or increasing its basal transcriptional rate, leads to an irreversible bistable switch: the switch remains on even when the activating signal is removed. Hence, the stem cell can be manipulated to be self-renewing without the requirement of input signals. We also suggest tests that could discriminate between a variety of feedforward regulation architectures of the target genes by OCT4, SOX2, and NANOG.
Conflict of interest statement
Figures




References
-
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. - PubMed
-
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. - PubMed
-
- Loh Y-H, Wu Q, Chew J-L, Vega VB, Zhang W, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Gen. 2006;38:431–440. - PubMed
-
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003;115:281–292. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

