CD4 and MHC-I downregulation are conserved in primary HIV-1 Nef alleles from brain and lymphoid tissues, but Pak2 activation is highly variable
- PMID: 16979207
- PMCID: PMC1995023
- DOI: 10.1016/j.virol.2006.07.053
CD4 and MHC-I downregulation are conserved in primary HIV-1 Nef alleles from brain and lymphoid tissues, but Pak2 activation is highly variable
Abstract
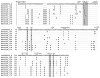
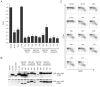
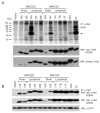
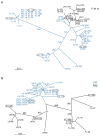
HIV-1 compartmentalization in the CNS has been demonstrated for gag, pol, and env genes. However, little is known about tissue compartmentalization of nef genes and their functional characteristics in brain. We have cloned 97 nef genes and characterized 10 Nef proteins from autopsy brain and lymphoid tissues from 2 patients with AIDS and HIV-1-associated dementia. Distinct compartmentalization of brain versus lymphoid nef genes was demonstrated within each patient. CD4 and MHC-I downregulation were conserved in all tissue-derived Nefs. However, MHC-I downregulation by brain-derived Nefs was weaker than downregulation by lymphoid-derived Nefs. The motifs KEEE- or EKEE- at the PACS-1 binding site represented brain-specific signature patterns in these 2 patients and contributed to the reduced MHC-I downregulation activity of brain-derived Nefs from these patients. Pak2 association was highly variable in Nefs from both patients. Three of 10 tissue-derived Nefs coimmunoprecipitated activated Pak2, with strong association demonstrated for only 2 Nefs. The ability of Nef to associate with activated Pak2 did not correlate with brain or lymphoid tissue origin. Nef genes from viruses isolated from brain by coculture with PBMC were not closely related to sequences amplified directly from brain tissue, suggesting that viral selection or adaptation occurred during coculture. This study of tissue-derived HIV-1 Nefs demonstrates that CD4 and MHC-I downregulation are highly conserved Nef functions, while Pak2 association is variable in late stage AIDS patients.
Figures


References
-
- Aiken C, Krause L, Chen YL, Trono D. Mutational analysis of HIV-1 Nef: identification of two mutants that are temperature-sensitive for CD4 downregulation. Virology. 1996;217(1):293–300. - PubMed
-
- Arold ST, Baur AS. Dynamic Nef and Nef dynamics: how structure could explain the complex activities of this small HIV protein. Trends Biochem Sci. 2001;26(6):356–63. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

