Structural basis of interaction between urokinase-type plasminogen activator and its receptor
- PMID: 16979660
- PMCID: PMC3443620
- DOI: 10.1016/j.jmb.2006.08.063
Structural basis of interaction between urokinase-type plasminogen activator and its receptor
Abstract
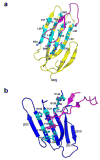
Recent studies indicate that binding of the urokinase-type plasminogen activator (uPA) to its high-affinity receptor (uPAR) orchestrates uPAR interactions with other cellular components that play a pivotal role in diverse (patho-)physiological processes, including wound healing, angiogenesis, inflammation, and cancer metastasis. However, notwithstanding the wealth of biochemical data available describing the activities of uPAR, little is known about the exact mode of uPAR/uPA interactions or the presumed conformational changes that accompany uPA/uPAR engagement. Here, we report the crystal structure of soluble urokinase plasminogen activator receptor (suPAR), which contains the three domains of the wild-type receptor but lacks the cell-surface anchoring sequence, in complex with the amino-terminal fragment of urokinase-type plasminogen activator (ATF), at the resolution of 2.8 A. We report the 1.9 A crystal structure of free ATF. Our results provide a structural basis, represented by conformational changes induced in uPAR, for several published biochemical observations describing the nature of uPAR/uPA interactions and provide insight into mechanisms that may be responsible for the cellular responses induced by uPA binding.
Figures




References
-
- Mondino A, Blasi F. uPA and uPAR in fibrinolysis, immunity and pathology. Trends Immunol. 2004;25:450–455. - PubMed
-
- Romer J, Nielsen BS, Ploug M. The urokinase receptor as a potential target in cancer therapy. Curr Pharm Des. 2004;10:2359–2376. - PubMed
-
- Rabbani SA, Mazar AP. The role of the plasminogen activation system in angiogenesis and metastasis. Surg Oncol Clin N Am. 2001;10:393–415. - PubMed
-
- Degryse B, Resnati M, Czekay RP, Loskutoff DJ, Blasi F. Domain 2 of the urokinase receptor contains an integrin-interacting epitope with intrinsic signaling activity: generation of a new integrin inhibitor. J Biol Chem. 2005;280:24792–24803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

