Large-scale gene expression differences across brain regions and inbred strains correlate with a behavioral phenotype
- PMID: 16980393
- PMCID: PMC1667050
- DOI: 10.1534/genetics.106.061481
Large-scale gene expression differences across brain regions and inbred strains correlate with a behavioral phenotype
Abstract
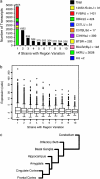
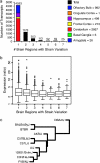
Behaviors are often highly heritable, polygenic traits. To investigate molecular mediators of behavior, we analyzed gene expression patterns across seven brain regions (amygdala, basal ganglia, cerebellum, frontal cortex, hippocampus, cingulate cortex, and olfactory bulb) of 10 different inbred mouse strains (129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BTBR T+ tf/J, C3H/HeJ, C57BL/6J, C57L/J, DBA/2J, and FVB/NJ). Extensive variation was observed across both strain and brain region. These data provide potential transcriptional intermediates linking polygenic variation to differences in behavior. For example, mice from different strains had variable performance on the rotarod task, which correlated with the expression of >2000 transcripts in the cerebellum. Correlation with this task was also found in the amygdala and hippocampus, but not in other regions examined, indicating the potential complexity of motor coordination. Thus we can begin to identify expression profiles contributing to behavioral phenotypes through variation in gene expression.
Figures

References
-
- Bast, T., and J. Feldon, 2003. Hippocampal modulation of sensorimotor processes. Prog. Neurobiol. 70: 319–345. - PubMed
-
- Benjamini, Y., and Y. Hochberg, 1995. Controlling the false discovery rate / a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57: 289–300.
-
- Bogue, M. A., and S. C. Grubb, 2004. The Mouse Phenome Project. Genetica 122: 71–74. - PubMed
-
- Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith et al., 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

