Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis
- PMID: 16980461
- PMCID: PMC1636309
- DOI: 10.1128/JB.00785-06
Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis
Abstract
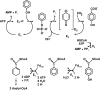
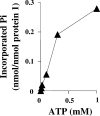
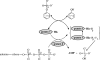
The anaerobic metabolism of phenol proceeds via carboxylation to 4-hydroxybenzoate by a two-step process involving seven proteins and two enzymes ("biological Kolbe-Schmitt carboxylation"). MgATP-dependent phosphorylation of phenol catalyzed by phenylphosphate synthase is followed by phenylphosphate carboxylation. Phenylphosphate synthase shows similarities to phosphoenolpyruvate (PEP) synthase and was studied for the bacterium Thauera aromatica. It consists of three proteins and transfers the beta-phosphoryl from ATP to phenol; the products are phenylphosphate, AMP, and phosphate. We showed that protein 1 becomes phosphorylated in the course of the reaction cycle by [beta-(32)P]ATP. This reaction requires protein 2 and is severalfold stimulated by protein 3. Stimulation of the reaction by 1 M sucrose is probably due to stabilization of the protein(s). Phosphorylated protein 1 transfers the phosphoryl group to phenolic substrates. The primary structure of protein 1 was analyzed by nanoelectrospray mass spectrometry after CNBr cleavage, trypsin digestion, and online high-pressure liquid chromatography at alkaline pH. His-569 was identified as the phosphorylated amino acid. We propose a catalytic ping-pong mechanism similar to that of PEP synthase. First, a diphosphoryl group is transferred to His-569 in protein 1, from which phosphate is cleaved to render the reaction unidirectional. Histidine phosphate subsequently serves as the actual phosphorylation agent.
Figures







Similar articles
-
Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica.J Bacteriol. 2004 Dec;186(23):8044-57. doi: 10.1128/JB.186.23.8044-8057.2004. J Bacteriol. 2004. PMID: 15547277 Free PMC article.
-
Genes involved in anaerobic metabolism of phenol in the bacterium Thauera aromatica.J Bacteriol. 2000 Oct;182(20):5849-63. doi: 10.1128/JB.182.20.5849-5863.2000. J Bacteriol. 2000. PMID: 11004186 Free PMC article.
-
Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica.J Bacteriol. 2004 Jul;186(14):4556-67. doi: 10.1128/JB.186.14.4556-4567.2004. J Bacteriol. 2004. PMID: 15231788 Free PMC article.
-
Anaerobic metabolism of phenol in proteobacteria and further studies of phenylphosphate carboxylase.Arch Microbiol. 2009 Dec;191(12):869-78. doi: 10.1007/s00203-009-0519-2. Epub 2009 Oct 17. Arch Microbiol. 2009. PMID: 19838679
-
Unusual reactions involved in anaerobic metabolism of phenolic compounds.Biol Chem. 2005 Oct;386(10):989-97. doi: 10.1515/BC.2005.115. Biol Chem. 2005. PMID: 16218871 Review.
Cited by
-
Anaerobic catabolism of aromatic compounds: a genetic and genomic view.Microbiol Mol Biol Rev. 2009 Mar;73(1):71-133. doi: 10.1128/MMBR.00021-08. Microbiol Mol Biol Rev. 2009. PMID: 19258534 Free PMC article. Review.
-
Anaerobic biodegradation of phenol in wastewater treatment: achievements and limits.Appl Microbiol Biotechnol. 2021 Mar;105(6):2195-2224. doi: 10.1007/s00253-021-11182-5. Epub 2021 Feb 25. Appl Microbiol Biotechnol. 2021. PMID: 33630152 Review.
-
Characterization of a two-component kinase that initiates the bacterial catabolism of hydroxyphenylethanones.J Biol Chem. 2025 Jun;301(6):110210. doi: 10.1016/j.jbc.2025.110210. Epub 2025 May 8. J Biol Chem. 2025. PMID: 40345584 Free PMC article.
-
Desulfatiglans anilini Initiates Degradation of Aniline With the Production of Phenylphosphoamidate and 4-Aminobenzoate as Intermediates Through Synthases and Carboxylases From Different Gene Clusters.Front Microbiol. 2020 Sep 4;11:2064. doi: 10.3389/fmicb.2020.02064. eCollection 2020. Front Microbiol. 2020. PMID: 33013754 Free PMC article.
-
Anaerobic metabolism of catechol by the denitrifying bacterium Thauera aromatica--a result of promiscuous enzymes and regulators?J Bacteriol. 2008 Mar;190(5):1620-30. doi: 10.1128/JB.01221-07. Epub 2007 Dec 21. J Bacteriol. 2008. PMID: 18156265 Free PMC article.
References
-
- Anders, H. J., A. Kaetzke, P. Kämpfer, W. Ludwig, and G. Fuchs. 1995. Taxonomic position aromatic-degrading denitrifying pseudomonad strains K 172 and KB 740 and their description as new members of the genera Thauera as Thauera aromatica sp. nov., and Azoarcus, as Azoarcus evansii sp. nov., respectively, members of the Proteobacteria. Int. J. Syst. Bacteriol. 45:327-333. - PubMed
-
- Aresta, M., and A. Dibenedetto. 2002. Development of environmentally friendly syntheses: use of enzymes and biomimetic systems for the direct carboxylation of organic substrates. J. Biotechnol. 90:113-128. - PubMed
-
- Berman, K. M., and M. Cohn. 1970. Phosphoenolpyruvate synthase of Escherichia coli: purification, some properties, and the role of divalent metal ions. J. Biol. Chem. 245:5309-5318. - PubMed
-
- Berman, K. M., and M. Cohn. 1970. Phosphoenolpyruvate synthase: partial reactions studied with adenosine triphosphate analogues and the inorganic phosphate-H218O exchange reaction. J. Biol. Chem. 245:5319-5325. - PubMed
-
- Besant, P. G., and P. V. Attwood. 1998. Problems with phosphoamino acid analysis using alkaline hydrolysis. Anal. Biochem. 265:187-190. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

