Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the phiKMV subgroup within the T7 supergroup
- PMID: 16980495
- PMCID: PMC1595506
- DOI: 10.1128/JB.00831-06
Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the phiKMV subgroup within the T7 supergroup
Abstract
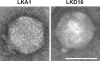
Lytic Pseudomonas aeruginosa phages LKD16 and LKA1 were locally isolated and morphologically classified as Podoviridae. While LKD16 adsorbs weakly to its host, LKA1 shows efficient adsorption (ka = 3.9 x 10(-9) ml min(-1)). LKA1, however, displays a narrow host range on clinical P. aeruginosa strains compared to LKD16. Genome analysis of LKD16 (43,200 bp) and LKA1 (41,593 bp) revealed that both phages have linear double-stranded DNA genomes with direct terminal repeats of 428 and 298 bp and encode 54 and 56 genes, respectively. The majority of the predicted structural proteins were experimentally confirmed as part of the phage particle using mass spectrometry. Phage LKD16 is closely related to bacteriophage phiKMV (83% overall DNA homology), allowing a more thoughtful gene annotation of both genomes. In contrast, LKA1 is more distantly related, lacking significant DNA homology and showing protein similarity to phiKMV in 48% of its gene products. The early region of the LKA1 genome has diverged strongly from phiKMV and LKD16, and intriguing differences in tail fiber genes of LKD16 and LKA1 likely reflect the observed discrepancy in infection-related properties. Nonetheless, general genome organization is clearly conserved among phiKMV, LKD16, and LKA1. The three phages carry a single-subunit RNA polymerase gene adjacent to the structural genome region, a feature which distinguishes them from other members of the T7 supergroup. Therefore, we propose that phiKMV represents an independent and widespread group of lytic P. aeruginosa phages within the T7 supergroup.
Figures

Similar articles
-
[Genome conservatism of phiKMV-like bacteriophages (T7 supergroup) active against Pseudomonas aeruginosa].Genetika. 2006 Jan;42(1):33-8. Genetika. 2006. PMID: 16523663 Russian.
-
Genomes of "phiKMV-like viruses" of Pseudomonas aeruginosa contain localized single-strand interruptions.Virology. 2009 Aug 15;391(1):1-4. doi: 10.1016/j.virol.2009.06.024. Epub 2009 Jul 9. Virology. 2009. PMID: 19592061
-
Characterization of a novel Pseudomonas aeruginosa bacteriophage, KPP25, of the family Podoviridae.Virus Res. 2014 Aug 30;189:43-6. doi: 10.1016/j.virusres.2014.04.019. Epub 2014 May 5. Virus Res. 2014. PMID: 24801109
-
Genomics of staphylococcal Twort-like phages--potential therapeutics of the post-antibiotic era.Adv Virus Res. 2012;83:143-216. doi: 10.1016/B978-0-12-394438-2.00005-0. Adv Virus Res. 2012. PMID: 22748811 Review.
-
Phage phiKZ-The First of Giants.Viruses. 2021 Jan 20;13(2):149. doi: 10.3390/v13020149. Viruses. 2021. PMID: 33498475 Free PMC article. Review.
Cited by
-
Complete genomic sequence of bacteriophage felix o1.Viruses. 2010 Mar;2(3):710-730. doi: 10.3390/v2030710. Epub 2010 Mar 9. Viruses. 2010. PMID: 21994654 Free PMC article.
-
Characterization of bacteriophages infecting clinical isolates of Pseudomonas aeruginosa stored in a culture collection.Braz J Med Biol Res. 2013 Aug;46(8):689-95. doi: 10.1590/1414-431X20132796. Epub 2013 Aug 13. Braz J Med Biol Res. 2013. PMID: 23969975 Free PMC article.
-
Isolation and characterization of a T7-like lytic phage for Pseudomonas fluorescens.BMC Biotechnol. 2008 Oct 27;8:80. doi: 10.1186/1472-6750-8-80. BMC Biotechnol. 2008. PMID: 18954452 Free PMC article.
-
Genomic Characterization of Sixteen Yersinia enterocolitica-Infecting Podoviruses of Pig Origin.Viruses. 2018 Apr 3;10(4):174. doi: 10.3390/v10040174. Viruses. 2018. PMID: 29614052 Free PMC article.
-
Integrative omics analysis of Pseudomonas aeruginosa virus PA5oct highlights the molecular complexity of jumbo phages.Environ Microbiol. 2020 Jun;22(6):2165-2181. doi: 10.1111/1462-2920.14979. Epub 2020 Mar 25. Environ Microbiol. 2020. PMID: 32154616 Free PMC article.
References
-
- Adams, M. H. 1959. Bacteriophages, p. 14-15. Interscience Publishers, Inc., New York, N.Y.
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. - PubMed
-
- Barenboim, M., C. Y. Chang, F. dib Hajj, and R. Young. 1999. Characterization of the dual start motif of a class II holin gene. Mol. Microbiol. 32:715-727. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

