The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants
- PMID: 16980538
- PMCID: PMC1626614
- DOI: 10.1105/tpc.106.045088
The chromatin assembly factor subunit FASCIATA1 is involved in homologous recombination in plants
Abstract
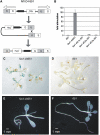
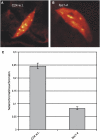
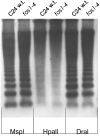
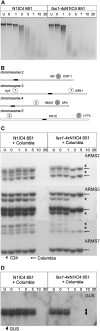
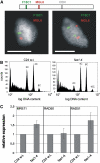
DNA replication in cycling eukaryotic cells necessitates the reestablishment of chromatin after nucleosome redistribution from the parental to the two daughter DNA strands. Chromatin assembly factor 1 (CAF-1), a heterotrimeric complex consisting of three subunits (p150/p60/p48), is one of the replication-coupled assembly factors involved in the reconstitution of S-phase chromatin. CAF-1 is required in vitro for nucleosome assembly onto newly replicated chromatin in human cells and Arabidopsis thaliana, and defects in yeast (Saccharomyces cerevisiae) affect DNA damage repair processes, predominantly those involved in genome stability. However, in vivo chromatin defects of caf-1 mutants in higher eukaryotes are poorly characterized. Here, we show that fasciata1-4 (fas1-4), a new allele of the Arabidopsis fas1 mutant defective in the p150 subunit of CAF-1, has a severe developmental phenotype, reduced heterochromatin content, and a more open conformation of euchromatin. Most importantly, homologous recombination (HR), a process involved in maintaining genome stability, is increased dramatically in fas1-4, as indicated by a 96-fold stimulation of intrachromosomal HR. Together with the open conformation of chromatin and the nearly normal expression levels of HR genes in the mutant, this result suggests that chromatin is a major factor restricting HR in plants.
Figures

References
-
- Adkins, M.W., and Tyler, J.K. (2004). The histone chaperone Asf1p mediates global chromatin disassembly in vivo. J. Biol. Chem. 279 52069–52074. - PubMed
-
- Alexeev, A., Mazin, A., and Kowalczykowski, S.C. (2003). Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10 182–186. - PubMed
-
- Barow, M., and Meister, A. (2003). Endopolyploidy in seed plants is differently correlated to systematics, organ, life strategy and genome size. Plant Cell Environ. 26 571–584.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

