Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals
- PMID: 16980559
- PMCID: PMC1630745
- DOI: 10.1104/pp.106.087676
Branching genes are conserved across species. Genes controlling a novel signal in pea are coregulated by other long-distance signals
Abstract
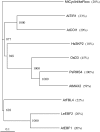
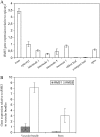
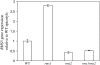
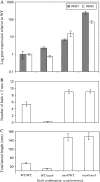
Physiological and genetic studies with the ramosus (rms) mutants in garden pea (Pisum sativum) and more axillary shoots (max) mutants in Arabidopsis (Arabidopsis thaliana) have shown that shoot branching is regulated by a network of long-distance signals. Orthologous genes RMS1 and MAX4 control the synthesis of a novel graft-transmissible branching signal that may be a carotenoid derivative and acts as a branching inhibitor. In this study, we demonstrate further conservation of the branching control system by showing that MAX2 and MAX3 are orthologous to RMS4 and RMS5, respectively. This is consistent with the long-standing hypothesis that branching in pea is regulated by a novel long-distance signal produced by RMS1 and RMS5 and that RMS4 is implicated in the response to this signal. We examine RMS5 expression and show that it is more highly expressed relative to RMS1, but under similar transcriptional regulation as RMS1. Further expression studies support the hypothesis that RMS4 functions in shoot and rootstock and participates in the feedback regulation of RMS1 and RMS5 expression. This feedback involves a second novel long-distance signal that is lacking in rms2 mutants. RMS1 and RMS5 are also independently regulated by indole-3-acetic acid. RMS1, rather than RMS5, appears to be a key regulator of the branching inhibitor. This study presents new interactions between RMS genes and provides further evidence toward the ongoing elucidation of a model of axillary bud outgrowth in pea.
Figures




References
-
- Aloni R, Schwalm K, Langhans M, Ullrich CI (2003) Gradual shifts in sites of free-auxin production during leaf-primordium development and their role in vascular differentiation and leaf morphogenesis in Arabidopsis. Planta 216: 841–853 - PubMed
-
- Arumingtyas EL, Floyd RS, Gregory MJ, Murfet IC (1992) Branching in Pisum: inheritance and allelism in tests with 17 ramosus mutants. Pisum Genet 24: 17–31
-
- Auldridge M, Block A, Vogel J, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty D, Klee H (2006) Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45: 982–993 - PubMed
-
- Bainbridge K, Sorefan K, Ward S, Leyser O (2005) Hormonally controlled expression of the Arabidopsis MAX4 shoot branching regulatory gene. Plant J 44: 569–580 - PubMed
-
- Bennett T, Sieberer T, Willett B, Booker J, Lusching C, Leyser O (2006) The Arabidopsis MAX pathway controls shoot branching by regulating auxin transport. Curr Biol 16: 553–563 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

