The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions
- PMID: 16980959
- PMCID: PMC2829982
- DOI: 10.1038/ncb1477
The chaperonin TRiC controls polyglutamine aggregation and toxicity through subunit-specific interactions
Abstract
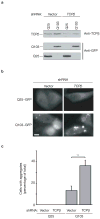
Misfolding and aggregation of proteins containing expanded polyglutamine repeats underlie Huntington's disease and other neurodegenerative disorders. Here, we show that the hetero-oligomeric chaperonin TRiC (also known as CCT) physically interacts with polyglutamine-expanded variants of huntingtin (Htt) and effectively inhibits their aggregation. Depletion of TRiC enhances polyglutamine aggregation in yeast and mammalian cells. Conversely, overexpression of a single TRiC subunit, CCT1, is sufficient to remodel Htt-aggregate morphology in vivo and in vitro, and reduces Htt-induced toxicity in neuronal cells. Because TRiC acts during de novo protein biogenesis, this chaperonin may have an early role preventing Htt access to pathogenic conformations. Based on the specificity of the Htt-CCT1 interaction, the CCT1 substrate-binding domain may provide a versatile scaffold for therapeutic inhibitors of neurodegenerative disease.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures




References
-
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. - PubMed
-
- Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. - PubMed
-
- Soto C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nature Rev Neurosci. 2003;4:49–60. - PubMed
-
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nature Rev Neurosci. 2005;6:11–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

