Binding of human SLBP on the 3'-UTR of histone precursor H4-12 mRNA induces structural rearrangements that enable U7 snRNA anchoring
- PMID: 16982637
- PMCID: PMC1635294
- DOI: 10.1093/nar/gkl666
Binding of human SLBP on the 3'-UTR of histone precursor H4-12 mRNA induces structural rearrangements that enable U7 snRNA anchoring
Abstract
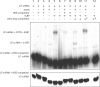
In metazoans, cell-cycle-dependent histones are produced from poly(A)-lacking mRNAs. The 3' end of histone mRNAs is formed by an endonucleolytic cleavage of longer precursors between a conserved stem-loop structure and a purine-rich histone downstream element (HDE). The cleavage requires at least two trans-acting factors: the stem-loop binding protein (SLBP), which binds to the stem-loop and the U7 snRNP, which anchors to histone pre-mRNAs by annealing to the HDE. Using RNA structure-probing techniques, we determined the secondary structure of the 3'-untranslated region (3'-UTR) of mouse histone pre-mRNAs H4-12, H1t and H2a-614. Surprisingly, the HDE is embedded in hairpin structures and is therefore not easily accessible for U7 snRNP anchoring. Probing of the 3'-UTR in complex with SLBP revealed structural rearrangements leading to an overall opening of the structure especially at the level of the HDE. Electrophoretic mobility shift assays demonstrated that the SLBP-induced opening of HDE actually facilitates U7 snRNA anchoring on the histone H4-12 pre-mRNAs 3' end. These results suggest that initial binding of the SLBP functions in making the HDE more accessible for U7 snRNA anchoring.
Figures



Similar articles
-
The polyribosomal protein bound to the 3' end of histone mRNA can function in histone pre-mRNA processing.Nucleic Acids Symp Ser. 1995;(33):234-6. Nucleic Acids Symp Ser. 1995. PMID: 8643381
-
U7 snRNP is recruited to histone pre-mRNA in a FLASH-dependent manner by two separate regions of the stem-loop binding protein.RNA. 2017 Jun;23(6):938-951. doi: 10.1261/rna.060806.117. Epub 2017 Mar 13. RNA. 2017. PMID: 28289156 Free PMC article.
-
Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility.Nature. 1990 Aug 16;346(6285):665-8. doi: 10.1038/346665a0. Nature. 1990. PMID: 1696685
-
Formation of the 3' end of histone mRNA.Gene. 1999 Oct 18;239(1):1-14. doi: 10.1016/s0378-1119(99)00367-4. Gene. 1999. PMID: 10571029 Review.
-
U2 snRNP: not just for poly(A) mRNAs.Mol Cell. 2007 Nov 9;28(3):353-4. doi: 10.1016/j.molcel.2007.10.015. Mol Cell. 2007. PMID: 17996698 Review.
Cited by
-
Three proteins of the U7-specific Sm ring function as the molecular ruler to determine the site of 3'-end processing in mammalian histone pre-mRNA.Mol Cell Biol. 2009 Aug;29(15):4045-56. doi: 10.1128/MCB.00296-09. Epub 2009 May 26. Mol Cell Biol. 2009. PMID: 19470752 Free PMC article.
-
U7 snRNAs: a computational survey.Genomics Proteomics Bioinformatics. 2007 Dec;5(3-4):187-95. doi: 10.1016/S1672-0229(08)60006-6. Genomics Proteomics Bioinformatics. 2007. PMID: 18267300 Free PMC article.
-
Secondary structure of the SARS-CoV-2 5'-UTR.RNA Biol. 2021 Apr;18(4):447-456. doi: 10.1080/15476286.2020.1814556. Epub 2020 Sep 23. RNA Biol. 2021. PMID: 32965173 Free PMC article.
-
RNA-binding protein GLD-1/quaking genetically interacts with the mir-35 and the let-7 miRNA pathways in Caenorhabditis elegans.Open Biol. 2013 Nov 20;3(11):130151. doi: 10.1098/rsob.130151. Open Biol. 2013. PMID: 24258276 Free PMC article.
-
Maternally encoded stem-loop-binding protein is degraded in 2-cell mouse embryos by the co-ordinated activity of two separately regulated pathways.Dev Biol. 2009 Apr 1;328(1):140-7. doi: 10.1016/j.ydbio.2009.01.018. Epub 2009 Jan 23. Dev Biol. 2009. PMID: 19298784 Free PMC article.
References
-
- Huynen M., Konings D., Hogeweg P. Equal G and C contents in histone genes indicate selection pressures on mRNA secondary structure. J. Mol. Evol. 1992;34:280–291. - PubMed
-
- Marzluff W.F. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr. Opin. Cell Biol. 2005;17:274–280. - PubMed
-
- Jaeger S., Barends S., Giegé R., Eriani G., Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. - PubMed
-
- Wang Z.F., Whitfield M.L., Ingledue T.C., 3rd, Dominski Z., Marzluff W.F. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNAs processing. Genes Dev. 1996;10:3028–3040. - PubMed

