A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation
- PMID: 16982646
- PMCID: PMC1635246
- DOI: 10.1093/nar/gkl472
A set of nearest neighbor parameters for predicting the enthalpy change of RNA secondary structure formation
Abstract
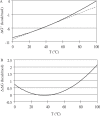
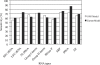
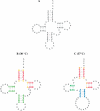
A complete set of nearest neighbor parameters to predict the enthalpy change of RNA secondary structure formation was derived. These parameters can be used with available free energy nearest neighbor parameters to extend the secondary structure prediction of RNA sequences to temperatures other than 37 degrees C. The parameters were tested by predicting the secondary structures of sequences with known secondary structure that are from organisms with known optimal growth temperatures. Compared with the previous set of enthalpy nearest neighbor parameters, the sensitivity of base pair prediction improved from 65.2 to 68.9% at optimal growth temperatures ranging from 10 to 60 degrees C. Base pair probabilities were predicted with a partition function and the positive predictive value of structure prediction is 90.4% when considering the base pairs in the lowest free energy structure with pairing probability of 0.99 or above. Moreover, a strong correlation is found between the predicted melting temperatures of RNA sequences and the optimal growth temperatures of the host organism. This indicates that organisms that live at higher temperatures have evolved RNA sequences with higher melting temperatures.
Figures
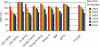


Similar articles
-
A sensitivity analysis of RNA folding nearest neighbor parameters identifies a subset of free energy parameters with the greatest impact on RNA secondary structure prediction.Nucleic Acids Res. 2017 Jun 2;45(10):6168-6176. doi: 10.1093/nar/gkx170. Nucleic Acids Res. 2017. PMID: 28334976 Free PMC article.
-
Thermodynamic characterization of single mismatches found in naturally occurring RNA.Biochemistry. 2007 Nov 20;46(46):13425-36. doi: 10.1021/bi701311c. Epub 2007 Oct 24. Biochemistry. 2007. PMID: 17958380
-
TurboFold: iterative probabilistic estimation of secondary structures for multiple RNA sequences.BMC Bioinformatics. 2011 Apr 20;12:108. doi: 10.1186/1471-2105-12-108. BMC Bioinformatics. 2011. PMID: 21507242 Free PMC article.
-
Revolutions in RNA secondary structure prediction.J Mol Biol. 2006 Jun 9;359(3):526-32. doi: 10.1016/j.jmb.2006.01.067. Epub 2006 Feb 6. J Mol Biol. 2006. PMID: 16500677 Review.
-
Measuring the thermodynamics of RNA secondary structure formation.Biopolymers. 1997;44(3):309-19. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. Biopolymers. 1997. PMID: 9591481 Review.
Cited by
-
Thermodynamic characterization of RNA 2 × 3 nucleotide internal loops.Biochemistry. 2012 Jul 3;51(26):5359-68. doi: 10.1021/bi3001227. Epub 2012 Jun 21. Biochemistry. 2012. PMID: 22720720 Free PMC article.
-
A Polymer Physics Framework for the Entropy of Arbitrary Pseudoknots.Biophys J. 2019 Aug 6;117(3):520-532. doi: 10.1016/j.bpj.2019.06.037. Epub 2019 Jul 10. Biophys J. 2019. PMID: 31353036 Free PMC article.
-
Improving RNA nearest neighbor parameters for helices by going beyond the two-state model.Nucleic Acids Res. 2018 Jun 1;46(10):4883-4892. doi: 10.1093/nar/gky270. Nucleic Acids Res. 2018. PMID: 29718397 Free PMC article.
-
Single-molecule mechanical unfolding and folding of a pseudoknot in human telomerase RNA.RNA. 2007 Dec;13(12):2175-88. doi: 10.1261/rna.676707. Epub 2007 Oct 24. RNA. 2007. PMID: 17959928 Free PMC article.
-
OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics.Nucleic Acids Res. 2008 Jul 1;36(Web Server issue):W104-8. doi: 10.1093/nar/gkn250. Epub 2008 May 19. Nucleic Acids Res. 2008. PMID: 18490376 Free PMC article.
References
-
- Nelson P., Kiriakidou M., Sharma A., Maniataki E., Mourelatos Z. The microRNA world: small is mighty. Trends Biochem. Sci. 2003;28:534–540. - PubMed
-
- Doudna J., Cech T. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. - PubMed
-
- Walter P., Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature. 1982;299:691–698. - PubMed
-
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. - PubMed
-
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. - PubMed

