Role of RNA polymerase III transcription factors in the selection of integration sites by the dictyostelium non-long terminal repeat retrotransposon TRE5-A
- PMID: 16982688
- PMCID: PMC1636787
- DOI: 10.1128/MCB.01348-06
Role of RNA polymerase III transcription factors in the selection of integration sites by the dictyostelium non-long terminal repeat retrotransposon TRE5-A
Abstract
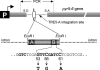
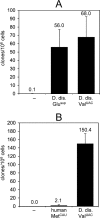
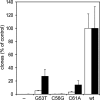
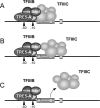
In the compact Dictyostelium discoideum genome, non-long terminal repeat (non-LTR) retrotransposons known as TREs avoid accidental integration-mediated gene disruption by targeting the vicinity of tRNA genes. In this study we provide the first evidence that proteins of a non-LTR retrotransposon interact with a target-specific transcription factor to direct its integration. We applied an in vivo selection system that allows for the isolation of natural TRE5-A integrations into a known genomic location upstream of tRNA genes. TRE5-A frequently modified the integration site in a way characteristic of other non-LTR retrotransposons by adding nontemplated extra nucleotides and generating small and extended target site deletions. Mutations within the B-box promoter of the targeted tRNA genes interfered with both the in vitro binding of RNA polymerase III transcription factor TFIIIC and the ability of TRE5-A to target these genes. An isolated B box was sufficient to enhance TRE5-A integration in the absence of a surrounding tRNA gene. The RNA polymerase III-transcribed ribosomal 5S gene recruits TFIIIC in a B-box-independent manner, yet it was readily targeted by TRE5-A in our assay. These results suggest a direct role of an RNA polymerase III transcription factor in the targeting process.
Figures






Similar articles
-
TRE5-A retrotransposition profiling reveals putative RNA polymerase III transcription complex binding sites on the Dictyostelium extrachromosomal rDNA element.PLoS One. 2017 Apr 13;12(4):e0175729. doi: 10.1371/journal.pone.0175729. eCollection 2017. PLoS One. 2017. PMID: 28406973 Free PMC article.
-
Transfer RNA gene-targeted retrotransposition of Dictyostelium TRE5-A into a chromosomal UMP synthase gene trap.J Mol Biol. 2002 Apr 26;318(2):273-85. doi: 10.1016/S0022-2836(02)00097-9. J Mol Biol. 2002. PMID: 12051837
-
Protein interactions involved in tRNA gene-specific integration of Dictyostelium discoideum non-long terminal repeat retrotransposon TRE5-A.Mol Cell Biol. 2007 Dec;27(24):8492-501. doi: 10.1128/MCB.01173-07. Epub 2007 Oct 8. Mol Cell Biol. 2007. PMID: 17923679 Free PMC article.
-
Transfer RNA gene-targeted integration: an adaptation of retrotransposable elements to survive in the compact Dictyostelium discoideum genome.Cytogenet Genome Res. 2005;110(1-4):288-98. doi: 10.1159/000084961. Cytogenet Genome Res. 2005. PMID: 16093681 Review.
-
Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark.Gene. 2012 Feb 10;493(2):169-75. doi: 10.1016/j.gene.2011.09.018. Epub 2011 Oct 1. Gene. 2012. PMID: 21986035 Review.
Cited by
-
Dictyostelium transfer RNA gene-targeting retrotransposons: Studying mobile element-host interactions in a compact genome.Mob Genet Elements. 2011 Jul;1(2):145-150. doi: 10.4161/mge.1.2.17369. Epub 2011 Jul 1. Mob Genet Elements. 2011. PMID: 22016864 Free PMC article.
-
TRE5-A retrotransposition profiling reveals putative RNA polymerase III transcription complex binding sites on the Dictyostelium extrachromosomal rDNA element.PLoS One. 2017 Apr 13;12(4):e0175729. doi: 10.1371/journal.pone.0175729. eCollection 2017. PLoS One. 2017. PMID: 28406973 Free PMC article.
-
Experimental annotation of the human pathogen Candida albicans coding and noncoding transcribed regions using high-resolution tiling arrays.Genome Biol. 2010;11(7):R71. doi: 10.1186/gb-2010-11-7-r71. Epub 2010 Jul 9. Genome Biol. 2010. PMID: 20618945 Free PMC article.
-
Convergent evolution of tRNA gene targeting preferences in compact genomes.Mob DNA. 2016 Aug 31;7(1):17. doi: 10.1186/s13100-016-0073-9. eCollection 2016. Mob DNA. 2016. PMID: 27583033 Free PMC article.
-
Suffix-specific RNAi leads to silencing of F element in Drosophila melanogaster.PLoS One. 2007 May 30;2(5):e476. doi: 10.1371/journal.pone.0000476. PLoS One. 2007. PMID: 17534426 Free PMC article.
References
-
- Beck, P., T. Dingermann, and T. Winckler. 2002. Transfer RNA gene-targeted retrotransposition of Dictyostelium TRE5-A into a chromosomal UMP synthase gene trap. J. Mol. Biol. 318:273-285. - PubMed
-
- Boeke, J. D., and S. E. Devine. 1998. Yeast retrotransposons: finding a nice quiet neighborhood. Cell 93:1087-1089. - PubMed
-
- Bukenberger, M., T. Dingermann, W. Meissner, K. H. Seifart, and T. Winckler. 1994. Isolation of transcription factor IIIC from Dictyostelium discoideum. Eur. J. Biochem. 220:839-846. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
