Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene
- PMID: 16982689
- PMCID: PMC1636772
- DOI: 10.1128/MCB.00678-06
Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene
Abstract
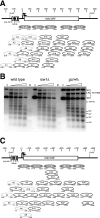
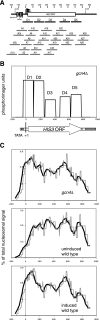
The effects of transcriptional activation on the chromatin structure of the Saccharomyces cerevisiae HIS3 gene were addressed by mapping the precise positions of nucleosomes in uninduced and induced chromatin. In the absence of the Gcn4p activator, the HIS3 gene is organized into a predominant nucleosomal array. In wild-type chromatin, this array is disrupted, and several alternative overlapping nucleosomal arrays are formed. The disruption of the predominant array also requires the SWI/SNF remodeling machine, indicating that the SWI/SNF complex plays an important role in nucleosome mobilization over the entire HIS3 gene. The Isw1 remodeling complex plays a more subtle role in determining nucleosome positions on HIS3, favoring positions different from those preferred by the SWI/SNF complex. Both the SWI/SNF and Isw1 complexes are constitutively present in HIS3 chromatin, although Isw1 tends to be excluded from the HIS3 promoter. Despite the apparent disorder of HIS3 chromatin generated by the formation of multiple nucleosomal arrays, nucleosome density profiles indicate that some long-range order is always present. We propose that Gcn4p stimulates nucleosome mobilization over the entire HIS3 gene by the SWI/SNF complex. We suggest that the net effect of interplay among remodeling machines at HIS3 is to create a highly dynamic chromatin structure.
Figures






Similar articles
-
Tup1-Ssn6 and Swi-Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene.Nucleic Acids Res. 2007;35(16):5520-31. doi: 10.1093/nar/gkm573. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704134 Free PMC article.
-
Antagonistic remodelling by Swi-Snf and Tup1-Ssn6 of an extensive chromatin region forms the background for FLO1 gene regulation.EMBO J. 2001 Sep 17;20(18):5219-31. doi: 10.1093/emboj/20.18.5219. EMBO J. 2001. PMID: 11566885 Free PMC article.
-
SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin.Proc Natl Acad Sci U S A. 2002 Nov 26;99(24):15381-6. doi: 10.1073/pnas.242536699. Epub 2002 Nov 13. Proc Natl Acad Sci U S A. 2002. PMID: 12432091 Free PMC article.
-
Nucleosome organization and targeting of SWI/SNF chromatin-remodeling complexes: contributions of the DNA sequence.Biochem Cell Biol. 2007 Aug;85(4):419-25. doi: 10.1139/O07-070. Biochem Cell Biol. 2007. PMID: 17713577 Review.
-
Remodeling chromatin structures for transcription: what happens to the histones?Bioessays. 1996 Nov;18(11):875-84. doi: 10.1002/bies.950181106. Bioessays. 1996. PMID: 8939065 Review.
Cited by
-
The yeast genome is globally accessible in living cells.Nat Struct Mol Biol. 2025 Feb;32(2):247-256. doi: 10.1038/s41594-024-01318-2. Epub 2024 Nov 25. Nat Struct Mol Biol. 2025. PMID: 39587299 Free PMC article.
-
Chromatin disassembly from the PHO5 promoter is essential for the recruitment of the general transcription machinery and coactivators.Mol Cell Biol. 2007 Sep;27(18):6372-82. doi: 10.1128/MCB.00981-07. Epub 2007 Jul 9. Mol Cell Biol. 2007. PMID: 17620413 Free PMC article.
-
Activator-dependent recruitment of SWI/SNF and INO80 during INO1 activation.Biochem Biophys Res Commun. 2008 Sep 5;373(4):602-6. doi: 10.1016/j.bbrc.2008.06.079. Epub 2008 Jun 30. Biochem Biophys Res Commun. 2008. PMID: 18593569 Free PMC article.
-
Gcn4 Binding in Coding Regions Can Activate Internal and Canonical 5' Promoters in Yeast.Mol Cell. 2018 Apr 19;70(2):297-311.e4. doi: 10.1016/j.molcel.2018.03.007. Epub 2018 Apr 5. Mol Cell. 2018. PMID: 29628310 Free PMC article.
-
Tup1-Ssn6 and Swi-Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene.Nucleic Acids Res. 2007;35(16):5520-31. doi: 10.1093/nar/gkm573. Epub 2007 Aug 17. Nucleic Acids Res. 2007. PMID: 17704134 Free PMC article.
References
-
- Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Top. Genet. Dev. 15:185-190. - PubMed
-
- Eriksson, P. R., G. Mendiratta, N. B. McLaughlin, T. G. Wolfsberg, L. Mariño-Ramírez, T. A. Pompa, M. Jainerin, D. Landsman, C.-H. Shen, and D. J. Clark. 2005. Global regulation by the yeast Spt10 protein is mediated through chromatin structure and the histone upstream activating sequence elements. Mol. Cell. Biol. 25:9127-9137. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
