The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3
- PMID: 16983073
- PMCID: PMC1600011
- DOI: 10.1073/pnas.0606385103
The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3
Abstract
In Arabidopsis thaliana, the promotion of flowering by cold temperatures, vernalization, is regulated via a floral-repressive MADS box transcription factor, FLOWERING LOCUS C (FLC). Vernalization leads to the epigenetic repression of FLC expression, a process that requires the polycomb group (PcG) protein VERNALIZATION 2 (VRN2) and the plant homeodomain protein VERNALIZATION INSENSITIVE 3 (VIN3). We demonstrate that the repression of FLC by vernalization requires homologues of other Polycomb Repressive Complex 2 proteins and VRN2. We show in planta that VRN2 and VIN3 are part of a large protein complex that can include the PcG proteins FERTILIZATION INDEPENDENT ENDOSPERM, CURLY LEAF, and SWINGER. These findings suggest a single protein complex is responsible for histone deacetylation at FLC and histone methylation at FLC in vernalized plants. The abundance of the complex increases during vernalization and declines after plants are returned to higher temperatures, consistent with the complex having a role in establishing FLC repression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



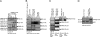
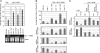
References
-
- Sheldon CC, Finnegan EJ, Dennis ES, Peacock WJ. Plant J. 2006;45:871–883. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

