Potential regulatory relationship between the nested gene DDC8 and its host gene tissue inhibitor of metalloproteinase-2
- PMID: 16985004
- PMCID: PMC3880020
- DOI: 10.1152/physiolgenomics.00160.2006
Potential regulatory relationship between the nested gene DDC8 and its host gene tissue inhibitor of metalloproteinase-2
Abstract
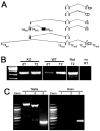
Nested genes are fairly common within the mammalian nervous system, yet few studies have examined whether the guest and host genes might be coordinately regulated. Tissue inhibitors of metalloproteinase (TIMPs) inhibit extracellular matrix proteolysis mediated by metzincin proteases. TIMP-2 is the only TIMP not nested within a synapsin gene. It does, however, serve as a host for differential display clone 8 (DDC8), a testis-specific gene whose expression is upregulated during spermatogenesis. Here, we demonstrate that DDC8 is not testis specific. Furthermore, DDC8 expression in nonneural and neural tissues mimics that of TIMP-2, including its upregulation in response to traumatic brain injury, suggesting a potential regulatory relationship. The most striking observation is that the TIMP-2 knockout mouse brain contains TIMP-2 mRNA encoding exons 2-5, which are downstream of DDC8, but not exon 1, which contains the signal sequence and cysteine residue required for MMP inhibition, indicating a functional knockout. That TIMP-2 transcripts in wild-type brain contain DDC8 sequence suggests alternative splicing between the two genes.
Figures




Similar articles
-
Expression and in-situ localization of genes coding for extracellular matrix proteins and extracellular matrix degrading proteases in pancreatic cancer.Int J Cancer. 1995 Aug 9;62(4):407-13. doi: 10.1002/ijc.2910620409. Int J Cancer. 1995. PMID: 7635566
-
Individual Timp deficiencies differentially impact pro-MMP-2 activation.J Biol Chem. 2006 Apr 14;281(15):10337-46. doi: 10.1074/jbc.M512009200. Epub 2006 Feb 9. J Biol Chem. 2006. PMID: 16469749
-
Sexually dimorphic diet-induced insulin resistance in obese tissue inhibitor of metalloproteinase-2 (TIMP-2)-deficient mice.Endocrinology. 2011 Apr;152(4):1300-13. doi: 10.1210/en.2010-1029. Epub 2011 Feb 1. Endocrinology. 2011. PMID: 21285317 Free PMC article.
-
Epilysin, a novel human matrix metalloproteinase (MMP-28) expressed in testis and keratinocytes and in response to injury.J Biol Chem. 2001 Mar 30;276(13):10134-44. doi: 10.1074/jbc.M001599200. Epub 2000 Dec 19. J Biol Chem. 2001. PMID: 11121398
-
Regulation of mRNAs encoding MMP-9 and MMP-2, and their inhibitors TIMP-1 and TIMP-2 by androgens in the rat ventral prostate.Mol Cell Endocrinol. 2008 Nov 6;294(1-2):10-8. doi: 10.1016/j.mce.2008.07.003. Epub 2008 Jul 15. Mol Cell Endocrinol. 2008. PMID: 18675881
Cited by
-
Preksha Dhyāna meditation induces alterations at the transcriptome level in novice and healthy college students.Saudi J Biol Sci. 2022 Apr;29(4):2299-2305. doi: 10.1016/j.sjbs.2021.11.060. Epub 2021 Dec 3. Saudi J Biol Sci. 2022. PMID: 35531197 Free PMC article.
-
Reflections on the evolution of the vertebrate tissue inhibitors of metalloproteinases.FASEB J. 2019 Jan;33(1):71-87. doi: 10.1096/fj.201801262R. Epub 2018 Aug 20. FASEB J. 2019. PMID: 30125136 Free PMC article. Review.
-
Secreted tissue inhibitor of matrix metalloproteinase restricts trans-synaptic signaling to coordinate synaptogenesis.J Cell Sci. 2017 Jul 15;130(14):2344-2358. doi: 10.1242/jcs.200808. Epub 2017 Jun 2. J Cell Sci. 2017. PMID: 28576972 Free PMC article.
-
Molecular mechanisms of tissue inhibitor of metalloproteinase 2 in the tumor microenvironment.Mol Cell Ther. 2014 Jun 3;2:17. doi: 10.1186/2052-8426-2-17. eCollection 2014. Mol Cell Ther. 2014. PMID: 26056585 Free PMC article. Review.
-
Tissue inhibitors of metalloproteinases in cell signaling: metalloproteinase-independent biological activities.Sci Signal. 2008 Jul 8;1(27):re6. doi: 10.1126/scisignal.127re6. Sci Signal. 2008. PMID: 18612141 Free PMC article. Review.
References
-
- Ahuja S, Ahuja P, Caffe AR, Ekstrom P, Abrahamson M, van Veen T. rd1 mouse retina shows imbalance in cellular distribution and levels of TIMP-1/MMP-9, TIMP-2/MMP-2 and sulfated glycosaminoglycans. Ophthalmic Res. 2006;38:125–136. - PubMed
-
- Akahane T, Akahane M, Shah A, Connor CM, Thorgeirsson UP. TIMP-1 inhibits microvascular endothelial cell migration by MMP-dependent and MMP-independent mechanisms. Exp Cell Res. 2004;310:158–167. - PubMed
-
- Baker AH, Edwards DR, Murphy G. Metalloproteinase inhibitors: biological actions and therapeutic opportunities. J Cell Sci. 2002;115:3719–3727. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

