Characterization of White bream virus reveals a novel genetic cluster of nidoviruses
- PMID: 16987966
- PMCID: PMC1642614
- DOI: 10.1128/JVI.01758-06
Characterization of White bream virus reveals a novel genetic cluster of nidoviruses
Abstract
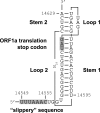
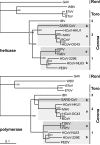
The order Nidovirales comprises viruses from the families Coronaviridae (genera Coronavirus and Torovirus), Roniviridae (genus Okavirus), and Arteriviridae (genus Arterivirus). In this study, we characterized White bream virus (WBV), a bacilliform plus-strand RNA virus isolated from fish. Analysis of the nucleotide sequence, organization, and expression of the 26.6-kb genome provided conclusive evidence for a phylogenetic relationship between WBV and nidoviruses. The polycistronic genome of WBV contains five open reading frames (ORFs), called ORF1a, -1b, -2, -3, and -4. In WBV-infected cells, three subgenomic RNAs expressing the structural proteins S, M, and N were identified. The subgenomic RNAs were revealed to share a 42-nucleotide, 5' leader sequence that is identical to the 5'-terminal genome sequence. The data suggest that a conserved nonanucleotide sequence, CA(G/A)CACUAC, located downstream of the leader and upstream of the structural protein genes acts as the core transcription-regulating sequence element in WBV. Like other nidoviruses with large genomes (>26 kb), WBV encodes in its ORF1b an extensive set of enzymes, including putative polymerase, helicase, ribose methyltransferase, exoribonuclease, and endoribonuclease activities. ORF1a encodes several membrane domains, a putative ADP-ribose 1"-phosphatase, and a chymotrypsin-like serine protease whose activity was established in this study. Comparative sequence analysis revealed that WBV represents a separate cluster of nidoviruses that significantly diverged from toroviruses and, even more, from coronaviruses, roniviruses, and arteriviruses. The study adds to the amazing diversity of nidoviruses and appeals for a more extensive characterization of nonmammalian nidoviruses to better understand the evolution of these largest known RNA viruses.
Figures






References
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
-
- Barrette-Ng, I. H., K. K. Ng, B. L. Mark, D. Van Aken, M. M. Cherney, C. Garen, Y. Kolodenko, A. E. Gorbalenya, E. J. Snijder, and M. N. James. 2002. Structure of arterivirus nsp4. The smallest chymotrypsin-like proteinase with an alpha/beta C-terminal extension and alternate conformations of the oxyanion hole. J. Biol. Chem. 277:39960-39966. - PubMed
-
- Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

