Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes
- PMID: 16987977
- PMCID: PMC1642607
- DOI: 10.1128/JVI.01414-06
Comparison of avian and human influenza A viruses reveals a mutational bias on the viral genomes
Abstract
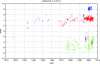
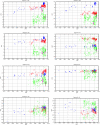
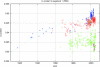
In the last few years, the genomic sequence data for thousands of influenza A virus strains, including the 1918 pandemic strain, and hundreds of isolates of the avian influenza virus H5N1, which is causing an increasing number of human fatalities, have become publicly available. This large quantity of sequence data allows us to do comparative genomics with the human and avian versions of the virus. We find that the nucleotide compositions of influenza A viruses infecting the two hosts are sufficiently different that we can determine the host at almost 100% accuracy. This assignment works at the segment level, which allows us to construct the reassortment history of individual segments within each strain. We suggest that the different nucleotide compositions can be explained by a host-dependent mutation bias. To support this idea, we estimate the fixation rates for the different polymerase segments and the ratios of synonymous to nonsynonymous changes. Additionally, we provide evidence supporting the hypothesis that the H1N1 influenza virus entered the human population just prior to the 1918 outbreak, with an earliest bound of 1910.
Figures
References
-
- Taubenberger, J. K., A. H. Reid, R. M. Lourens, R. Wang, G. Jin, and T. G. Fanning. 2005. Characterization of the 1918 influenza virus polymerase genes. Nature 437:889-893. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

