Characterization of igaB, a second immunoglobulin A1 protease gene in nontypeable Haemophilus influenzae
- PMID: 16988265
- PMCID: PMC1594874
- DOI: 10.1128/IAI.00796-06
Characterization of igaB, a second immunoglobulin A1 protease gene in nontypeable Haemophilus influenzae
Abstract
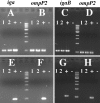
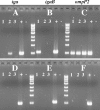
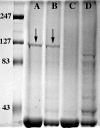
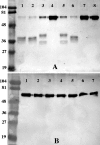
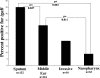
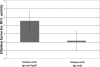
Nontypeable Haemophilus influenzae is an important respiratory pathogen, causing otitis media in children and lower respiratory tract infection in adults with chronic obstructive pulmonary disease (COPD). Immunoglobulin A1 (IgA1) protease is a well-described protein and potential virulence factor in this organism as well as other respiratory pathogens. IgA1 proteases cleave human IgA1, are involved in invasion, and display immunomodulatory effects. We have identified a second IgA1 protease gene, igaB, in H. influenzae that is present in addition to the previously described IgA1 protease gene, iga. Reverse transcriptase PCR and IgA1 protease assays indicated that the gene is transcribed, expressed, and enzymatically active in H. influenzae. The product of this gene is a type 2 IgA1 protease with homology to the iga gene of Neisseria species. Mutants that were deficient in iga, igaB, and both genes were constructed in H. influenzae strain 11P6H, a strain isolated from a patient with COPD who was experiencing an exacerbation. Analysis of these mutants indicated that igaB is the primary mediator of IgA1 protease activity in this strain. IgA1 protease activity assays on 20 clinical isolates indicated that the igaB gene is associated with increased levels of IgA1 protease activity. Approximately one-third of 297 strains of H. influenzae of diverse clinical and geographic origin contained igaB. Significant differences in the prevalence of igaB were observed among isolates from different sites of isolation (sputum > middle ear > nasopharynx). These data support the hypothesis that the newly discovered igaB gene is a potential virulence factor in nontypeable H. influenzae.
Figures





Similar articles
-
Expression of IgA Proteases by Haemophilus influenzae in the Respiratory Tract of Adults With Chronic Obstructive Pulmonary Disease.J Infect Dis. 2015 Dec 1;212(11):1798-805. doi: 10.1093/infdis/jiv299. Epub 2015 May 20. J Infect Dis. 2015. PMID: 25995193 Free PMC article.
-
A clonal group of nontypeable Haemophilus influenzae with two IgA proteases is adapted to infection in chronic obstructive pulmonary disease.PLoS One. 2011;6(10):e25923. doi: 10.1371/journal.pone.0025923. Epub 2011 Oct 5. PLoS One. 2011. PMID: 21998721 Free PMC article.
-
Nontypeable Haemophilus influenzae in carriage and disease: a difference in IgA1 protease activity levels.JAMA. 2002 Apr 3;287(13):1699-705. doi: 10.1001/jama.287.13.1699. JAMA. 2002. PMID: 11926894
-
IgA1 protease.Int J Biochem Cell Biol. 2006;38(8):1244-8. doi: 10.1016/j.biocel.2005.10.005. Epub 2005 Nov 2. Int J Biochem Cell Biol. 2006. PMID: 16293440 Free PMC article. Review.
-
Antigenic and genetic heterogeneity among Haemophilus influenzae and Neisseria IgA1 proteases.Adv Exp Med Biol. 1995;371A:599-603. doi: 10.1007/978-1-4615-1941-6_126. Adv Exp Med Biol. 1995. PMID: 8525998 Review. No abstract available.
Cited by
-
Specific IgA and metalloproteinase activity in bronchial secretions from stable chronic obstructive pulmonary disease patients colonized by Haemophilus influenzae.Respir Res. 2012 Dec 11;13(1):113. doi: 10.1186/1465-9921-13-113. Respir Res. 2012. PMID: 23228114 Free PMC article.
-
Expression of IgA Proteases by Haemophilus influenzae in the Respiratory Tract of Adults With Chronic Obstructive Pulmonary Disease.J Infect Dis. 2015 Dec 1;212(11):1798-805. doi: 10.1093/infdis/jiv299. Epub 2015 May 20. J Infect Dis. 2015. PMID: 25995193 Free PMC article.
-
Mycobacterium tuberculosis serine protease Rv3668c can manipulate the host-pathogen interaction via Erk-NF-κB axis-mediated cytokine differential expression.J Interferon Cytokine Res. 2014 Sep;34(9):686-98. doi: 10.1089/jir.2013.0071. Epub 2014 Mar 31. J Interferon Cytokine Res. 2014. PMID: 24684623 Free PMC article.
-
Non-typeable Haemophilus influenzae invasion and persistence in the human respiratory tract.Front Cell Infect Microbiol. 2011 Nov 18;1:1. doi: 10.3389/fcimb.2011.00001. eCollection 2011. Front Cell Infect Microbiol. 2011. PMID: 22919570 Free PMC article. Review.
-
Immunoglobulin A Protease Variants Facilitate Intracellular Survival in Epithelial Cells By Nontypeable Haemophilus influenzae That Persist in the Human Respiratory Tract in Chronic Obstructive Pulmonary Disease.J Infect Dis. 2017 Dec 5;216(10):1295-1302. doi: 10.1093/infdis/jix471. J Infect Dis. 2017. PMID: 28968876 Free PMC article.
References
-
- Ayala, B. P., B. Vasquez, S. Clary, J. A. Tainer, K. Rodland, and M. So. 2001. The pilus-induced Ca2+ flux triggers lysosome exocytosis and increases the amount of Lamp1 accessible to Neisseria IgA1 protease. Cell Microbiol. 3:265-275. - PubMed
-
- Beck, S. C., and T. F. Meyer. 2000. IgA1 protease from Neisseria gonorrhoeae inhibits TNF-α-mediated apoptosis of human monocytic cells. FEBS Lett. 472:287-292. - PubMed
-
- Brenner, D. J., L. W. Mayer, G. M. Carlone, L. H. Harrison, W. F. Bibb, M. C. Brandileone, F. O. Sottnek, K. Irino, M. W. Reeves, J. M. Swenson, et al. 1988. Biochemical, genetic, and epidemiologic characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazilian purpuric fever. J. Clin. Microbiol. 26:1524-1534. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

