Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits
- PMID: 16990592
- PMCID: PMC1785147
- DOI: 10.1182/blood-2006-07-038232
Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits
Abstract
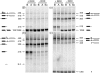
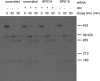
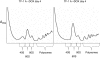
Diamond-Blackfan anemia (DBA) typically presents with red blood cell aplasia that usually manifests in the first year of life. The only gene currently known to be mutated in DBA encodes ribosomal protein S19 (RPS19). Previous studies have shown that the yeast RPS19 protein is required for a specific step in the maturation of 40S ribosomal subunits. Our objective here was to determine whether the human RPS19 protein functions at a similar step in 40S subunit maturation. Studies where RPS19 expression is reduced by siRNA in the hematopoietic cell line, TF-1, show that human RPS19 is also required for a specific step in the maturation of 40S ribosomal subunits. This maturation defect can be monitored by studying rRNA-processing intermediates along the ribosome synthesis pathway. Analysis of these intermediates in CD34- cells from the bone marrow of patients with DBA harboring mutations in RPS19 revealed a pre-rRNA-processing defect similar to that observed in TF-1 cells where RPS19 expression was reduced. This defect was observed to a lesser extent in CD34+ cells from patients with DBA who have mutations in RPS19.
Figures
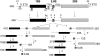

Similar articles
-
Impaired ribosome biogenesis in Diamond-Blackfan anemia.Blood. 2007 Feb 1;109(3):1275-83. doi: 10.1182/blood-2006-07-038372. Epub 2006 Oct 19. Blood. 2007. PMID: 17053056 Free PMC article.
-
Cells depleted for RPS19, a protein associated with Diamond Blackfan Anemia, show defects in 18S ribosomal RNA synthesis and small ribosomal subunit production.Blood Cells Mol Dis. 2007 Jul-Aug;39(1):35-43. doi: 10.1016/j.bcmd.2007.02.001. Epub 2007 Mar 21. Blood Cells Mol Dis. 2007. PMID: 17376718
-
Deficiency of ribosomal protein S19 in CD34+ cells generated by siRNA blocks erythroid development and mimics defects seen in Diamond-Blackfan anemia.Blood. 2005 Jun 15;105(12):4627-34. doi: 10.1182/blood-2004-08-3115. Epub 2004 Dec 30. Blood. 2005. PMID: 15626736
-
The functions of RPS19 and their relationship to Diamond-Blackfan anemia: a review.Mol Genet Metab. 2007 Apr;90(4):358-62. doi: 10.1016/j.ymgme.2006.11.004. Epub 2006 Dec 18. Mol Genet Metab. 2007. PMID: 17178250 Review.
-
Diamond Blackfan anemia: ribosomal proteins going rogue.Semin Hematol. 2011 Apr;48(2):89-96. doi: 10.1053/j.seminhematol.2011.02.005. Semin Hematol. 2011. PMID: 21435505 Review.
Cited by
-
A Review of Diamond-Blackfan Anemia: Current Evidence on Involved Genes and Treatment Modalities.Cureus. 2020 Aug 25;12(8):e10019. doi: 10.7759/cureus.10019. Cureus. 2020. PMID: 32983714 Free PMC article. Review.
-
Ribosomal and hematopoietic defects in induced pluripotent stem cells derived from Diamond Blackfan anemia patients.Blood. 2013 Aug 8;122(6):912-21. doi: 10.1182/blood-2013-01-478321. Epub 2013 Jun 6. Blood. 2013. PMID: 23744582 Free PMC article.
-
5'UTR variants of ribosomal protein S19 transcript determine translational efficiency: implications for Diamond-Blackfan anemia and tissue variability.PLoS One. 2011 Mar 11;6(3):e17672. doi: 10.1371/journal.pone.0017672. PLoS One. 2011. PMID: 21412415 Free PMC article.
-
The role of haploinsufficiency of RPS14 and p53 activation in the molecular pathogenesis of the 5q- syndrome.Pediatr Rep. 2011 Jun 22;3 Suppl 2(Suppl 2):e10. doi: 10.4081/pr.2011.s2.e10. Pediatr Rep. 2011. PMID: 22053272 Free PMC article.
-
Innate immune system activation in zebrafish and cellular models of Diamond Blackfan Anemia.Sci Rep. 2018 Mar 26;8(1):5165. doi: 10.1038/s41598-018-23561-6. Sci Rep. 2018. PMID: 29581525 Free PMC article.
References
-
- Bagby GC, Lipton JM, Sloand EM, Schiffer CA. Marrow failure. Hematology. 2004;2004:318–336. - PubMed
-
- Draptchinskaia N, Gustavsson P, Andersson B, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. - PubMed
-
- Gazda HT, Zhong R, Long L, et al. RNA and protein evidence for haplo-insufficiency in Diamond-Blackfan anaemia patients with RPS19 mutations. Br J Haematol. 2004;127:105–113. - PubMed
-
- Willig TN, Draptchinskaia N, Dianzani I, et al. Mutations in ribosomal protein S19 gene and Diamond Blackfan anemia: wide variations in phenotypic expression. Blood. 1999;94:4294–4306. - PubMed
-
- Liu JM, Ellis SR. Ribosomes and marrow failure: coincidental association or molecular paradigm? Blood. 2006;107:4583–4588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

