An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation
- PMID: 16990799
- PMCID: PMC1589998
- DOI: 10.1038/sj.emboj.7601339
An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation
Abstract
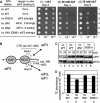
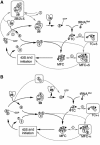
In eukaryotic translation initiation, the eIF2.GTP/Met-tRNA(i)(Met) ternary complex (TC) binds the eIF3/eIF1/eIF5 complex to form the multifactor complex (MFC), whereas eIF2.GDP binds the pentameric factor eIF2B for guanine nucleotide exchange. eIF5 and the eIF2Bvarepsilon catalytic subunit possess a conserved eIF2-binding site. Nearly half of cellular eIF2 forms a complex with eIF5 lacking Met-tRNA(i)(Met), and here we investigate its physiological significance. eIF5 overexpression increases the abundance of both eIF2/eIF5 and TC/eIF5 complexes, thereby impeding eIF2B reaction and MFC formation, respectively. eIF2Bvarepsilon mutations, but not other eIF2B mutations, enhance the ability of overexpressed eIF5 to compete for eIF2, indicating that interaction of eIF2Bvarepsilon with eIF2 normally disrupts eIF2/eIF5 interaction. Overexpression of the catalytic eIF2Bvarepsilon segment similarly exacerbates eIF5 mutant phenotypes, supporting the ability of eIF2Bvarepsilon to compete with MFC. Moreover, we show that eIF5 overexpression does not generate aberrant MFC lacking tRNA(i)(Met), suggesting that tRNA(i)(Met) is a vital component promoting MFC assembly. We propose that the eIF2/eIF5 complex represents a cytoplasmic reservoir for eIF2 that antagonizes eIF2B-promoted guanine nucleotide exchange, enabling coordinated regulation of translation initiation.
Figures





References
-
- Algire MA, Maag D, Lorsch JR (2005) Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 1–12 - PubMed
-
- Alone PV, Dever TE (2006) Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP–GTP exchange factors eIF5 and eIF2Beplsilon. J Biol Chem 281: 12636–12644 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

