Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis
- PMID: 16997968
- PMCID: PMC1698177
- DOI: 10.1128/JB.01213-06
Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis
Abstract
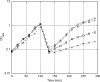
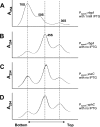
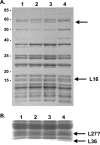
GTPases have been demonstrated to be necessary for the proper assembly of the ribosome in bacteria and eukaryotes. Here, we show that the essential GTPases YphC and YsxC are required for large ribosomal subunit biogenesis in Bacillus subtilis. Sucrose density gradient centrifugation of large ribosomal subunits isolated from YphC-depleted cells and YsxC-depleted cells indicates that they are similar to the 45S intermediate previously identified in RbgA-depleted cells. The sedimentation of the large-subunit intermediate isolated from YphC-depleted cells was identical to the intermediate found in RbgA-depleted cells, while the intermediate isolated from YsxC-depleted cells sedimented slightly slower than 45S, suggesting that it is a novel intermediate. Analysis of the protein composition of the large-subunit intermediates isolated from either YphC-depleted cells or YsxC-depleted cells indicated that L16 and L36 are missing. Purified YphC and YsxC are able to interact with the ribosome in vitro, supporting a direct role for these two proteins in the assembly of the 50S subunit. Our results indicate that, as has been demonstrated for Saccharomyces cerevisiae ribosome biogenesis, bacterial 50S ribosome assembly requires the function of multiple essential GTPases.
Figures


References
-
- Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. - PubMed
-
- Ban, N., P. Nissen, J. Hansen, P. B. Moore, and T. A. Steitz. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905-920. - PubMed
-
- Bashan, A., I. Agmon, R. Zarivach, F. Schluenzen, J. Harms, R. Berisio, H. Bartels, F. Franceschi, T. Auerbach, H. A. Hansen, E. Kossoy, M. Kessler, and A. Yonath. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol. Cell 11:91-102. - PubMed
-
- Britton, R. A., B. S. Powell, S. Dasgupta, Q. Sun, W. Margolin, J. R. Lupski, and D. L. Court. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27:739-750. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

