Nanoliter high throughput quantitative PCR
- PMID: 17000636
- PMCID: PMC1635282
- DOI: 10.1093/nar/gkl639
Nanoliter high throughput quantitative PCR
Abstract
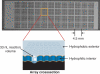
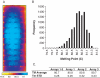
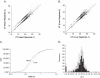
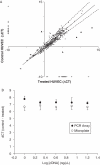
Understanding biological complexity arising from patterns of gene expression requires accurate and precise measurement of RNA levels across large numbers of genes simultaneously. Real time PCR (RT-PCR) in a microtiter plate is the preferred method for quantitative transcriptional analysis but scaling RT-PCR to higher throughputs in this fluidic format is intrinsically limited by cost and logistic considerations. Hybridization microarrays measure the transcription of many thousands of genes simultaneously yet are limited by low sensitivity, dynamic range, accuracy and sample throughput. The hybrid approach described here combines the superior accuracy, precision and dynamic range of RT-PCR with the parallelism of a microarray in an array of 3072 real time, 33 nl polymerase chain reactions (RT-PCRs) the size of a microscope slide. RT-PCR is demonstrated with an accuracy and precision equivalent to the same assay in a 384-well microplate but in a 64-fold smaller reaction volume, a 24-fold higher analytical throughput and a workflow compatible with standard microplate protocols.
Figures

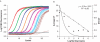

References
-
- Lipshutz R.J., Morris D., Chee M., Hubbell E., Kozal M.J., Shah N., Shen N., Yang R., Fodor S.P. Using oligonucleotide probe arrays to access genetic diversity. Biotechniques. 1995;19:442–447. - PubMed
-
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;5235:368–369. - PubMed
-
- Gershon D. Micorarray technology: an array of opportunites. Nature. 2002;416:885–891. - PubMed
-
- Murphy D. Gene expression studies using microarrays: principles, problems and prospects. Adv. Physiol. Educ. 2002;26:256–270. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

