Capturing genomic signatures of DNA sequence variation using a standard anonymous microarray platform
- PMID: 17000641
- PMCID: PMC1636412
- DOI: 10.1093/nar/gkl478
Capturing genomic signatures of DNA sequence variation using a standard anonymous microarray platform
Abstract
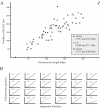
Comparative genomics, using the model organism approach, has provided powerful insights into the structure and evolution of whole genomes. Unfortunately, only a small fraction of Earth's biodiversity will have its genome sequenced in the foreseeable future. Most wild organisms have radically different life histories and evolutionary genomics than current model systems. A novel technique is needed to expand comparative genomics to a wider range of organisms. Here, we describe a novel approach using an anonymous DNA microarray platform that gathers genomic samples of sequence variation from any organism. Oligonucleotide probe sequences placed on a custom 44 K array were 25 bp long and designed using a simple set of criteria to maximize their complexity and dispersion in sequence probability space. Using whole genomic samples from three known genomes (mouse, rat and human) and one unknown (Gonystylus bancanus), we demonstrate and validate its power, reliability, transitivity and sensitivity. Using two separate statistical analyses, a large numbers of genomic 'indicator' probes were discovered. The construction of a genomic signature database based upon this technique would allow virtual comparisons and simple queries could generate optimal subsets of markers to be used in large-scale assays, using simple downstream techniques. Biologists from a wide range of fields, studying almost any organism, could efficiently perform genomic comparisons, at potentially any phylogenetic level after performing a small number of standardized DNA microarray hybridizations. Possibilities for refining and expanding the approach are discussed.
Figures


References
-
- Miller W., Makova K.D., Nekrutenko A., Hardison R.C. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. - PubMed
-
- NCBI. Genomic biology. National Center for Biotechnology Information. 2006 http://www.ncbi.nih.gov/Genomes
-
- Metzker M.L. Emerging technologies in DNA sequencing. Genome Res. 2005;15:1767–1776. - PubMed
-
- Shendure J., Mitra R.D., Varma C., Church G.M. Advanced sequencing technologies: methods and goals. Nature Rev. Genet. 2004;5:335–344. - PubMed

