Differential role of CbpA and PspA in modulation of in vitro CXC chemokine responses of respiratory epithelial cells to infection with Streptococcus pneumoniae
- PMID: 17000730
- PMCID: PMC1698053
- DOI: 10.1128/IAI.00954-06
Differential role of CbpA and PspA in modulation of in vitro CXC chemokine responses of respiratory epithelial cells to infection with Streptococcus pneumoniae
Abstract
Respiratory epithelial cells play an active part in the host response to respiratory pathogens, such as Streptococcus pneumoniae, by releasing chemokines responsible for neutrophil recruitment. In order to investigate the role of specific pneumococcal virulence factors in eliciting CXC chemokine responses, type II pneumocytes (A549) and nasopharyngeal cells (Detroit-562) were infected with S. pneumoniae D39 or mutants lacking choline-binding protein A (CbpA), pneumococcal surface protein A (PspA), or specific domains thereof. In response to wild-type D39, both A549 and Detroit-562 cells showed a significant increase in CXC chemokine mRNA and interleukin-8 protein. This response was increased twofold when a cbpA deletion mutant (DeltaCbpA) was used, suggesting that CbpA inhibits CXC chemokine induction. All three N-terminal domains of CbpA are required for this effect, as in-frame deletion of the respective region of cbpA had the same effect on the CXC chemokine response as deletion of cbpA altogether. Infection with a pspA deletion mutant (DeltaPspA) led to a twofold decrease in the CXC chemokine response of A549 but not Detroit-562 cells, compared to infection with D39 at 2 h. Thus, PspA appears to have the ability to stimulate early CXC chemokine release from A549 cells. Deletion of the region of pspA encoding the first N-terminal alpha-helical domain reduced the ability of S. pneumoniae to elicit a chemokine response to the same degree as deletion of pspA altogether. Thus, the N termini of CbpA and PspA exert differential effects on CXC chemokine induction in epithelial cells infected with S. pneumoniae.
Figures




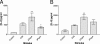


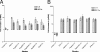
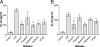
References
-
- Balachandran, P., A. Brooks-Walter, A. Virolainen-Julkunen, S. K. Hollingshead, and D. E. Briles. 2002. Role of pneumococcal surface protein C in nasopharyngeal carriage and pneumonia and its ability to elicit protection against carriage of Streptococcus pneumoniae. Infect. Immun. 70:2526-2534. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

