Mutational analysis of class A and class B penicillin-binding proteins in Streptococcus gordonii
- PMID: 17000738
- PMCID: PMC1693976
- DOI: 10.1128/AAC.00677-06
Mutational analysis of class A and class B penicillin-binding proteins in Streptococcus gordonii
Abstract
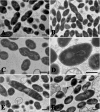
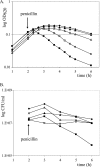
High-molecular-weight (HMW) penicillin-binding proteins (PBPs) are divided into class A and class B PBPs, which are bifunctional transpeptidases/transglycosylases and monofunctional transpeptidases, respectively. We determined the sequences for the HMW PBP genes of Streptococcus gordonii, a gingivo-dental commensal related to Streptococcus pneumoniae. Five HMW PBPs were identified, including three class A (PBPs 1A, 1B, and 2A) and two class B (PBPs 2B and 2X) PBPs, by homology with those of S. pneumoniae and by radiolabeling with [3H]penicillin. Single and double deletions of each of them were achieved by allelic replacement. All could be deleted, except for PBP 2X, which was essential. Morphological alterations occurred after deletion of PBP 1A (lozenge shape), PBP 2A (separation defect and chaining), and PBP 2B (aberrant septation and premature lysis) but not PBP 1B. The muropeptide cross-link patterns remained similar in all strains, indicating that cross-linkage for one missing PBP could be replaced by others. However, PBP 1A mutants presented shorter glycan chains (by 30%) and a relative decrease (25%) in one monomer stem peptide. Growth rate and viability under aeration, hyperosmolarity, and penicillin exposure were affected primarily in PBP 2B-deleted mutants. In contrast, chain-forming PBP 2A-deleted mutants withstood better aeration, probably because they formed clusters that impaired oxygen diffusion. Double deletion could be generated with any PBP combination and resulted in more-altered mutants. Thus, single deletion of four of the five HMW genes had a detectable effect on the bacterial morphology and/or physiology, and only PBP 1B seemed redundant a priori.
Figures



Similar articles
-
Promoter and transcription analysis of penicillin-binding protein genes in Streptococcus gordonii.Antimicrob Agents Chemother. 2007 Aug;51(8):2774-83. doi: 10.1128/AAC.01127-06. Epub 2007 May 14. Antimicrob Agents Chemother. 2007. PMID: 17502405 Free PMC article.
-
Functional Insights into the High-Molecular-Mass Penicillin-Binding Proteins of Streptococcus agalactiae Revealed by Gene Deletion and Transposon Mutagenesis Analysis.J Bacteriol. 2021 Aug 9;203(17):e0023421. doi: 10.1128/JB.00234-21. Epub 2021 Aug 9. J Bacteriol. 2021. PMID: 34124943 Free PMC article.
-
Mutations in penicillin-binding protein (PBP) genes and in non-PBP genes during selection of penicillin-resistant Streptococcus gordonii.Antimicrob Agents Chemother. 2006 Dec;50(12):4053-61. doi: 10.1128/AAC.00676-06. Epub 2006 Sep 25. Antimicrob Agents Chemother. 2006. PMID: 17000741 Free PMC article.
-
Streptococcus faecium mutants that are temperature sensitive for cell growth and show alterations in penicillin-binding proteins.J Bacteriol. 1987 Jun;169(6):2432-9. doi: 10.1128/jb.169.6.2432-2439.1987. J Bacteriol. 1987. PMID: 3584060 Free PMC article.
-
Penicillin-binding protein-mediated resistance in pneumococci and staphylococci.J Infect Dis. 1999 Mar;179 Suppl 2:S353-9. doi: 10.1086/513854. J Infect Dis. 1999. PMID: 10081507 Review.
Cited by
-
Promoter and transcription analysis of penicillin-binding protein genes in Streptococcus gordonii.Antimicrob Agents Chemother. 2007 Aug;51(8):2774-83. doi: 10.1128/AAC.01127-06. Epub 2007 May 14. Antimicrob Agents Chemother. 2007. PMID: 17502405 Free PMC article.
-
Fitness cost and impaired survival in penicillin-resistant Streptococcus gordonii isolates selected in the laboratory.Antimicrob Agents Chemother. 2008 Jan;52(1):337-9. doi: 10.1128/AAC.00939-07. Epub 2007 Nov 12. Antimicrob Agents Chemother. 2008. PMID: 17999969 Free PMC article.
-
Role of PBPD1 in stimulation of Listeria monocytogenes biofilm formation by subminimal inhibitory β-lactam concentrations.Antimicrob Agents Chemother. 2014 Nov;58(11):6508-17. doi: 10.1128/AAC.03671-14. Epub 2014 Aug 18. Antimicrob Agents Chemother. 2014. PMID: 25136010 Free PMC article.
-
Penicillin-Binding Protein 3 Is Essential for Growth of Pseudomonas aeruginosa.Antimicrob Agents Chemother. 2016 Dec 27;61(1):e01651-16. doi: 10.1128/AAC.01651-16. Print 2017 Jan. Antimicrob Agents Chemother. 2016. PMID: 27821444 Free PMC article.
-
Functional Insights into the High-Molecular-Mass Penicillin-Binding Proteins of Streptococcus agalactiae Revealed by Gene Deletion and Transposon Mutagenesis Analysis.J Bacteriol. 2021 Aug 9;203(17):e0023421. doi: 10.1128/JB.00234-21. Epub 2021 Aug 9. J Bacteriol. 2021. PMID: 34124943 Free PMC article.
References
-
- Amsterdam, D. 1996. Susceptibility testing of antimicrobials in liquid medium, p. 52-111. In V. Lorian (ed.), Antibiotics in laboratory medicine. Williams and Wilkins, Baltimore, Md.
-
- Bischoff, M., M. Roos, J. Putnik, A. Wada, P. Glanzmann, P. Giachino, P. Vaudaux, and B. Berger-Bachi. 2001. Involvement of multiple genetic loci in Staphylococcus aureus teicoplanin resistance. FEMS Microbiol. Lett. 194:77-82. - PubMed
-
- Bizzini, A., P. A. Majcherczyk, S. Beggah-Möller, B. Soldo, J. Entenza, M. Gaillard, P. Moreillon, and V. Lazarevic. Submitted for publication. - PubMed
-
- Cabeen, M. T., and C. Jacobs-Wagner. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601-610. - PubMed
-
- Chen, J.-D., and D. A. Morrison. 1988. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene 64:155-164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

