Upf1/Upf2 regulation of 3' untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay
- PMID: 17000771
- PMCID: PMC1636803
- DOI: 10.1128/MCB.02251-05
Upf1/Upf2 regulation of 3' untranslated region splice variants of AUF1 links nonsense-mediated and A+U-rich element-mediated mRNA decay
Abstract
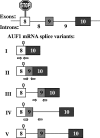
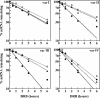
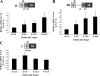
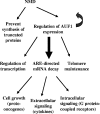
AUF1 is an RNA-binding protein that targets mRNAs containing A+U-rich elements (AREs) for rapid cytoplasmic turnover. Alternative pre-mRNA splicing produces five variants of AUF1 mRNA that differ in the composition of their 3'-untranslated regions (3'-UTRs). Previous work suggested that this heterogeneity in 3'-UTR sequence could regulate AUF1 expression by two potential mechanisms. First, AUF1 may regulate its own expression by binding to AREs in 3'-UTR splice variants that retain intron 9. The second potential mechanism, and the focus of this report, is regulation of a subset of 3'-UTR splice variants by the nonsense-mediated mRNA decay (NMD) pathway. Two of the five AUF1 mRNA 3'-UTR variants position the translational termination codon more than 50 nucleotides upstream of an exon-exon junction, creating a potential triggering signal for NMD in mammalian cells. Disruption of cellular NMD pathways by RNA interference-mediated knockdown of Upf1/Rent1 or Upf2/Rent2 or transfection of a dominant-negative Upf1 mutant specifically enhanced expression of these two candidate NMD substrate mRNAs in cells, involving stabilization of each transcript. Ribonucleoprotein immunoprecipitation experiments revealed that both Upf1 and Upf2 can associate with an NMD-sensitive AUF1 mRNA 3'-UTR variant in cells. Finally, quantitation of AUF1 mRNA 3'-UTR splice variants during murine embryonic development showed that the expression of NMD-sensitive AUF1 mRNAs is specifically enhanced as development proceeds, contributing to dynamic changes in AUF1 3'-UTR structures during embryogenesis. Together, these studies provide the first evidence of linkage between the nonsense- and ARE-mediated mRNA decay pathways, which may constitute a new mechanism regulating the expression of ARE-containing mRNAs.
Figures





References
-
- Bejerano, G., M. Pheasant, I. Makunin, S. Stephen, W. J. Kent, J. S. Mattick, and D. Haussler. 2004. Ultraconserved elements in the human genome. Science 304:1321-1325. - PubMed
-
- Boffelli, D., M. A. Nobrega, and E. M. Rubin. 2004. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 5:456-465. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
