Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine
- PMID: 17002302
- PMCID: PMC2580076
- DOI: 10.1021/bi061293x
Mechanistic investigations of the pseudouridine synthase RluA using RNA containing 5-fluorouridine
Abstract
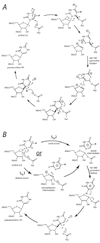
The pseuoduridine synthases (psi synthases) isomerize uridine (U) to pseudouridine (psi) in RNA, and they fall into five families that share very limited sequence similarity but have the same overall fold and active-site architecture, including an essential Asp. The mechanism by which the psi synthases operate remains unknown, and mechanistic work has largely made use of RNA containing 5-fluorouridine (f5U) in place of U. The psi synthase TruA forms a covalent adduct with such RNA, and heat disruption of the adduct generates a hydrated product of f5U, which was reasonably concluded to result from the hydrolysis of an ester linkage between the essential Asp and f5U. In contrast, the psi synthase TruB, which is a member of a different family, does not form an adduct with f5U in RNA but catalyzes the rearrangement and hydration of the f5U, which labeling studies with [18O]water showed does not result from ester hydrolysis. To extend the line of mechanistic investigation to another family of psi synthases and an enzyme that makes an adduct with f5U in RNA, the behavior of RluA toward RNA containing f5U was examined. Stem-loop RNAs are shown to be good substrates for RluA. Heat denaturation of the adduct between RluA and RNA containing f5U produces a hydrated nucleoside product, and labeling studies show that hydration does not occur by ester hydrolysis. These results are interpreted in light of a consistent mechanistic scheme for the handling of f5U by psi synthases.
Figures







Similar articles
-
The handling of the mechanistic probe 5-fluorouridine by the pseudouridine synthase TruA and its consistency with the handling of the same probe by the pseudouridine synthases TruB and RluA.Biochemistry. 2011 Jan 25;50(3):426-36. doi: 10.1021/bi101737z. Epub 2010 Dec 29. Biochemistry. 2011. PMID: 21142053 Free PMC article.
-
The pseudouridine synthases: revisiting a mechanism that seemed settled.J Am Chem Soc. 2004 Oct 13;126(40):12758-9. doi: 10.1021/ja046375s. J Am Chem Soc. 2004. PMID: 15469254
-
Not all pseudouridine synthases are potently inhibited by RNA containing 5-fluorouridine.RNA. 2004 Feb;10(2):192-9. doi: 10.1261/rna.5100104. RNA. 2004. PMID: 14730018 Free PMC article.
-
Pseudouridine in RNA: what, where, how, and why.IUBMB Life. 2000 May;49(5):341-51. doi: 10.1080/152165400410182. IUBMB Life. 2000. PMID: 10902565 Review.
-
Pseudouridine synthases.Chem Biol. 2006 Nov;13(11):1125-35. doi: 10.1016/j.chembiol.2006.09.009. Chem Biol. 2006. PMID: 17113994 Review.
Cited by
-
Unexpected linear ion trap collision-induced dissociation and Fourier transform ion cyclotron resonance infrared multi-photon dissociation fragmentation of a hydrated C-glycoside of 5-fluorouridine formed by the action of the pseudouridine synthases RluA and TruB.Rapid Commun Mass Spectrom. 2011 Sep 30;25(18):2627-32. doi: 10.1002/rcm.5162. Rapid Commun Mass Spectrom. 2011. PMID: 23657957 Free PMC article.
-
RNA-based 5-fluorouracil toxicity requires the pseudouridylation activity of Cbf5p.Genetics. 2008 May;179(1):323-30. doi: 10.1534/genetics.107.082727. Genetics. 2008. PMID: 18493057 Free PMC article.
-
The mechanism of pseudouridine synthases from a covalent complex with RNA, and alternate specificity for U2605 versus U2604 between close homologs.Nucleic Acids Res. 2014 Feb;42(3):2037-48. doi: 10.1093/nar/gkt1050. Epub 2013 Nov 7. Nucleic Acids Res. 2014. PMID: 24214967 Free PMC article.
-
Pseudouridine: still mysterious, but never a fake (uridine)!RNA Biol. 2014;11(12):1540-54. doi: 10.4161/15476286.2014.992278. RNA Biol. 2014. PMID: 25616362 Free PMC article. Review.
-
Pseudouridines of tRNA Anticodon Stem-Loop Have Unexpected Role in Mutagenesis in Pseudomonas sp.Microorganisms. 2020 Dec 23;9(1):25. doi: 10.3390/microorganisms9010025. Microorganisms. 2020. PMID: 33374637 Free PMC article.
References
-
- Grosjean H, Benne R. Modification and Editing of RNA. Washington, DC: ASM Press; 1998.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

