Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1
- PMID: 17002499
- PMCID: PMC1570379
- DOI: 10.1371/journal.pgen.0020155
Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1
Abstract
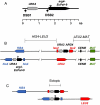
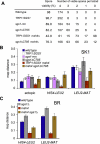
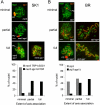
Sgs1, the budding yeast homolog of the mammalian BLM helicase, has been implicated in preventing excess recombination during both vegetative growth and meiosis. Most meiotic crossover (CO) recombination requires full function of a set of yeast proteins (Zip1, Zip2, Zip3, Zip4/Spo22, Mer3, Msh4, and Msh5, termed the SIC or ZMM proteins) that are also required for homologous chromosome synapsis. We report here genetic and molecular assays showing that sgs1 single mutants display relatively modest increases in CO recombination (less than 1.6-fold relative to wild-type). In contrast, a much greater CO increase is seen when an sgs1 mutation is introduced into the CO- and synapsis-deficient zip1, zip2, zip3, mer3, or msh4 mutants (2- to 8-fold increase). Furthermore, close juxtaposition of the axes of homologous chromosomes is restored. CO restoration in the mutants is not accompanied by significant changes in noncrossover (NCO) recombinant frequencies. These findings show that Sgs1 has potent meiotic anti-CO activity, which is normally antagonized by SIC/ZMM proteins. Our data reinforce previous proposals for an early separation of meiotic processes that form CO and NCO recombinants.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures

References
-
- Jones GH. Chiasmata. In: Moens PB, editor. Meiosis. Orlando (Florida): Academic Press; 1987. pp. 213–244.
-
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. - PubMed
-
- Petronczki M, Siomos MF, Nasmyth K. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

